Figure 8.
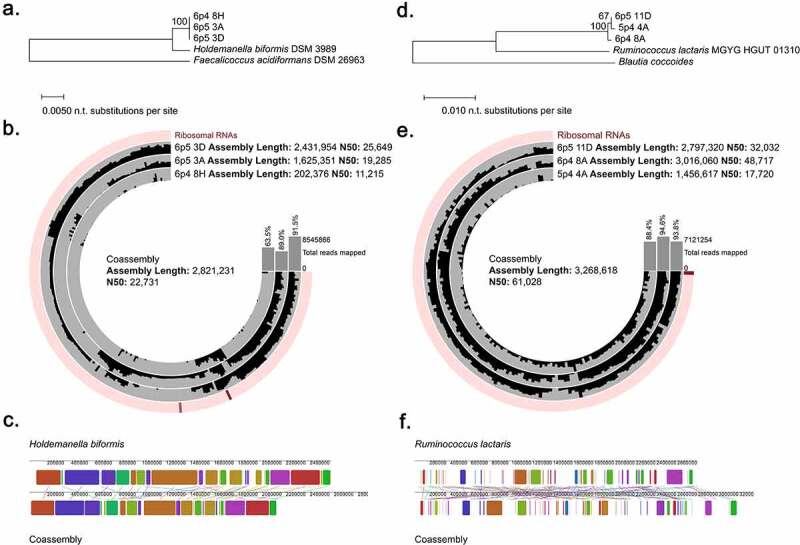
Co-assembly of similar rare microbes improves genomic assemblies and reveals novel taxa. (a) Phylogenetic tree based on full length 16S rRNA genes extracted from single cell assemblies of cells classified as Holdemanella by V4 amplicon sequencing. (b) Coverage plot of each individual cell mapped to the co-assembly. Assembly statistics for the co-assembly and individual cell assemblies are displayed as well as the percentage of total reads mapped the assembly. (c) Alignment of co-assembled genome to Holdemanella biformis genome. (d) Phylogenetic tree based on full length 16S rRNA genes extracted from single cell assemblies of cells classified as Ruminococcus lactaris by V4 amplicon sequencing. (e) Coverage plot of each individual cell mapped to the coassembly. Assembly statistics for the co-assembly and individual cell assemblies are displayed as well as the percentage of total reads mapped the assembly. (f) Alignment of co-assembled genome to Ruminococcus lactaris genome.
