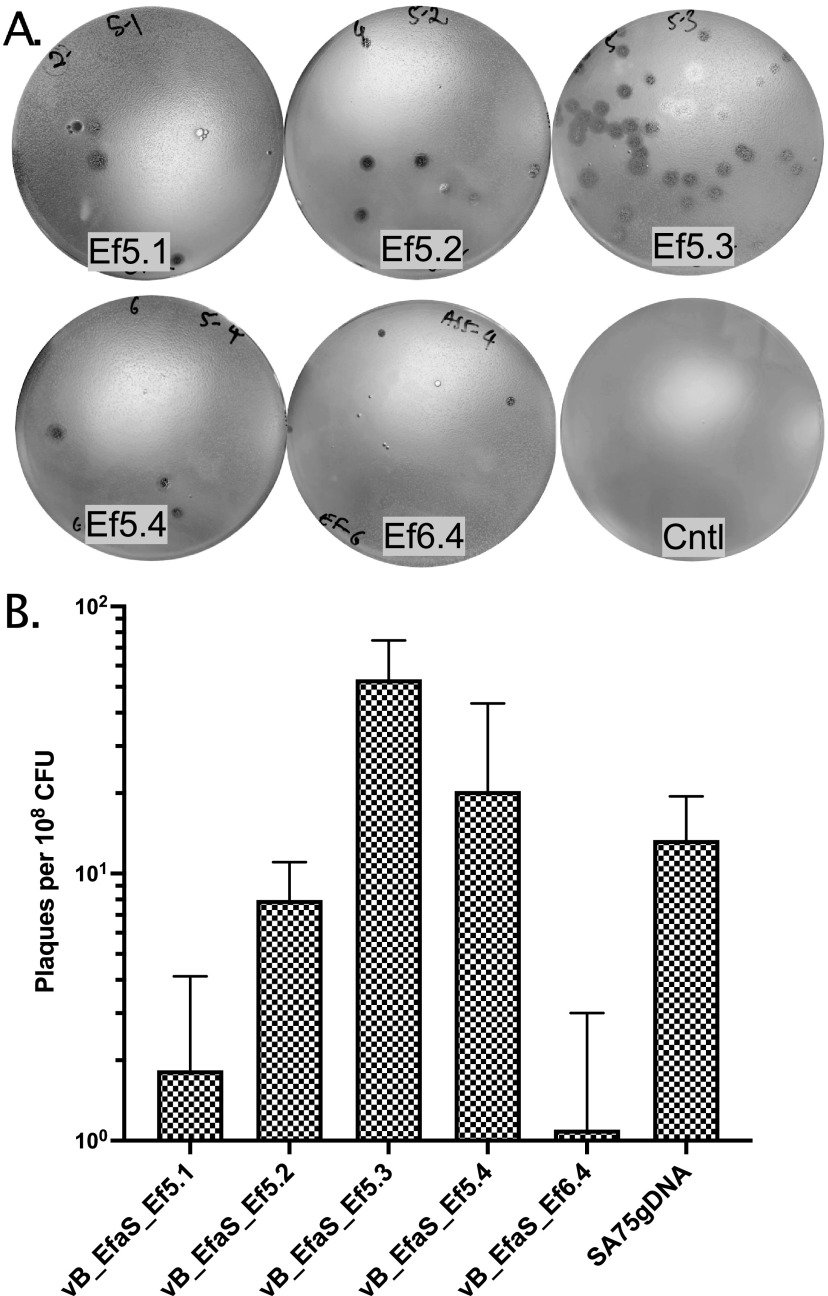
FIG 5.
Cross-genus transfection of E. faecalis phage genomic DNA using NEST. Appropriate amount (100 to 200 ng) of E. faecalis phage ɸ5.1, ɸ5.2, ɸ5.3, ɸ5.4 and ɸ6.4 gDNA was transformed into S. aureus cells (∼9 × 108 CFU) using the NEST method and incubated for 24 h at 23°C. (A) Transformants were mixed with the corresponding indicator strain (E. faecalis AH5 for ɸ5.1, ɸ5.2, ɸ5.3, and ɸ5.4, or AH6 for ɸ6.4) and plated on BHI media using the top agar overlay method and incubated at 30°C for 24 to 48 h to observe the plaques. The control plate (Cntl) was a lawn of S. aureus RN4220 cells mixed with E. faecalis AH5 lacking phage DNA. (B) The efficiency of boot-up was determined by calculating the number of plaques per 108 CFU of NEST competent cells of each E. faecalis phage and SA75 for comparison. Each data point represents the mean from three independent experiments, and the error bars indicate standard error.