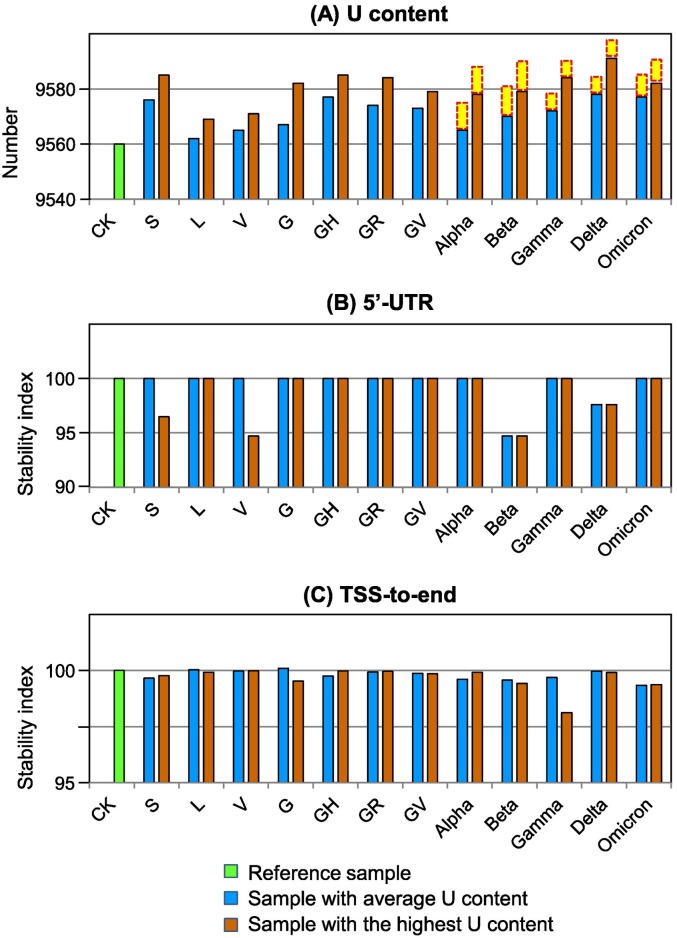
Fig. 4.
Genome stability of SARS-CoV-2. For genome stability comparison, two virus samples were taken for calculating free energy of their genomic segments. One of the samples has average U content, while the other has the highest U content. (A) U content in various samples. Yellow block on top of data bar indicates U number lost from deletions (as shown in Fig. 1) in the five variants. (B) and (C) Stability of 5′-UTR (untranslated region) and TSS (translation start site)-to-end region in various samples. Please refer to Table S4 for detailed sample information and free energy values. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Genome stability of SARS-CoV-2. For genome stability comparison, two virus samples were taken for calculating free energy of their genomic segments. One of the samples has average U content, while the other has the highest U content. (A) U content in various samples. Yellow block on top of data bar indicates U number lost from deletions (as shown in Fig. 1) in the five variants. (B) and (C) Stability of 5′-UTR (untranslated region) and TSS (translation start site)-to-end region in various samples. Please refer to Table S4 for detailed sample information and free energy values. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)