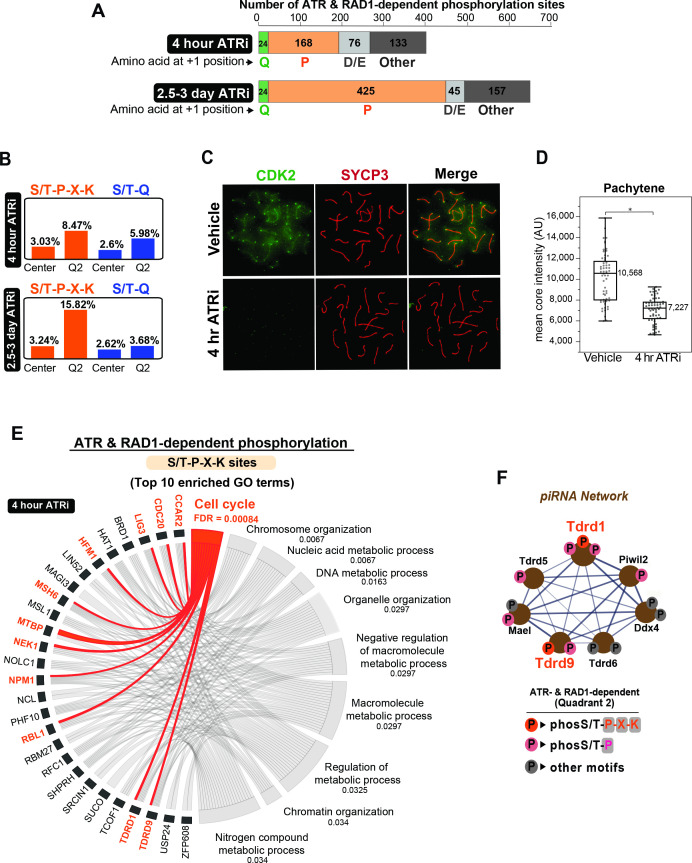
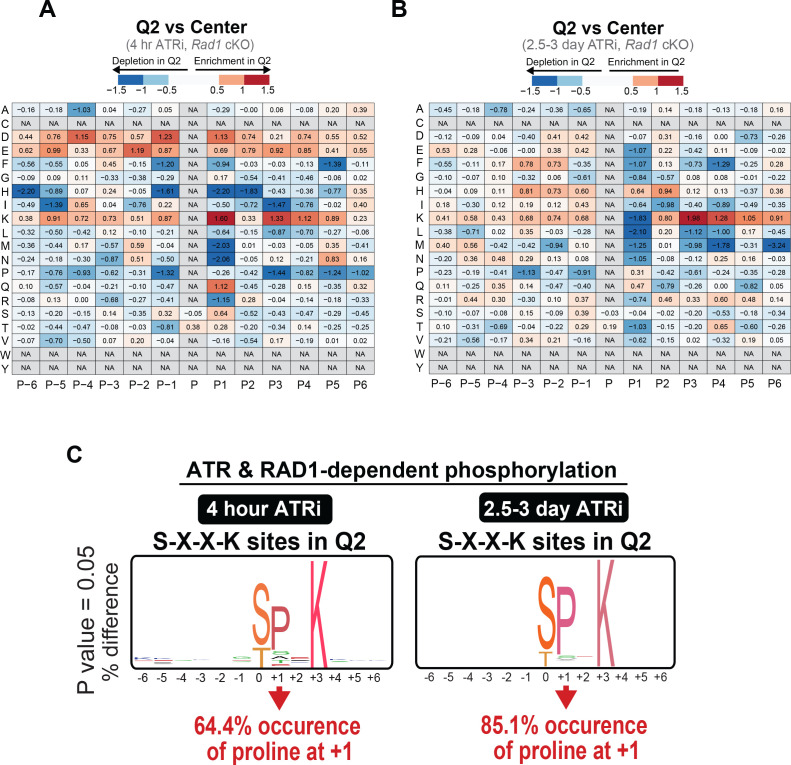
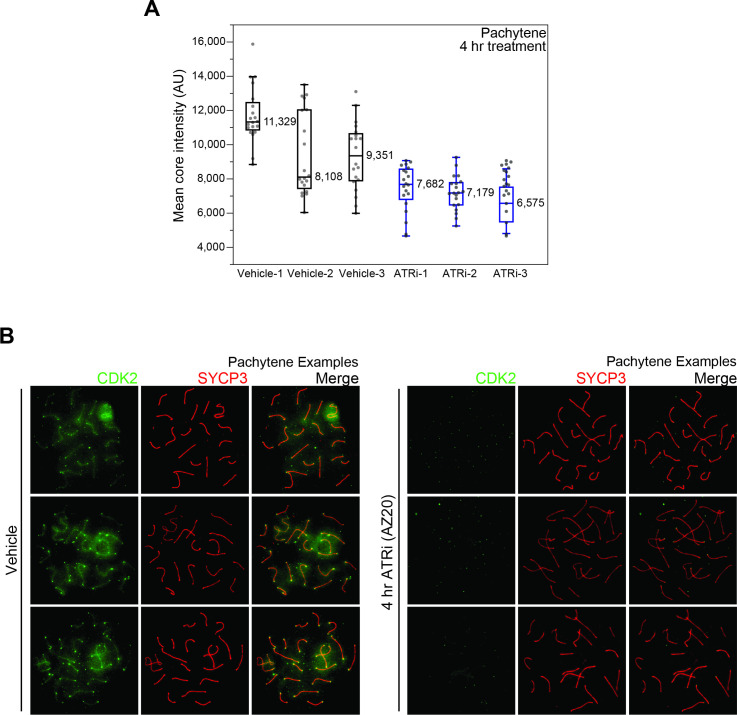
Figure 5. Enrichment of S/T-P-X-K phosphorylation motif in the set of ATR and RAD1-dependent signaling events.
(A) Bar graph depicting the count of Q2 phosphopeptides with the indicated amino acids at the +1 position. (B) Bar graph of the percentage of indicated phospho-motifs in the center (unchanged events) and Q2. (C) Immunofluorescence of meiotic spreads from mice treated with vehicle or 50 mg/kg AZ20 for 4 hr and stained for CDK2 (green) and SYCP3 (red). (D) Quantification of CDK2 signal intensity of pachytene-staged cells in (C) (three vehicle mice, n = 60 cells; three ATRi mice, n = 60 cells p=0.0368 measured by Student’s t-test) (see Materials and methods for more details). (E) Chord plot of gene ontology (GO) analysis of ATR and RAD1-dependent S/T-P-X-K phosphorylation events using STRING. The top 10 significantly enriched biological processes GO terms were selected. False discovery rate (FDR) for GO term enrichment is shown below each term. GO terms are shown on the right and proteins found for each term on the left. (F) STRING analysis cluster of piRNA-related proteins with Q2 phosphorylation.