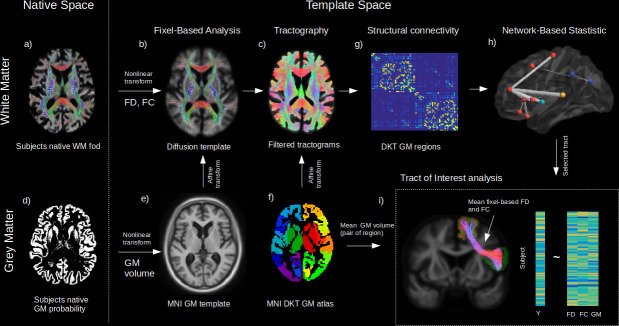
Figure 1. Method workflow.
The main steps of the methods are shown from the native space (left) to template space (right). For the white matter (WM) (upper panels), native diffusion-weighted MRIs were first preprocessed to obtain individual normalized WM FODs (a). WM FODs were non-linearly registered to a study-specific WM FOD template (b), to obtain the fibre density (FD), and fibre cross-section metrics (FC), later used in whole brain fixel-based analysis. The template space WM FODs were then used to generate individual probabilistic tractograms (c). For the grey matter (GM) (lower panels), native space GM probability maps were warped to a study specific GM template in MNI space to obtain individual template space GM volume (e). An affine transform was estimated between MNI template and the diffusion template space which was subsequently applied to the Desikan-Killiany (DKT) GM atlas to bring the DKT atlas in diffusion space (f). Individual structural connectivity matrices were then obtained by counting the amount of fibres connecting each pair of GM regions within the DKT atlas (g). Significant difference in connectivity for a given dependant variable (Y) was then tested using the network-based statistic enhanced (h). Significant predictors (connections) were selected to access the relative importance of GM volume and WM (FD and FC) within each connection in predicting Y (h), where mean FD and FC were obtained in fixels belonging to the connection streamlines and GM was the average of both GM regions volume for each subject.