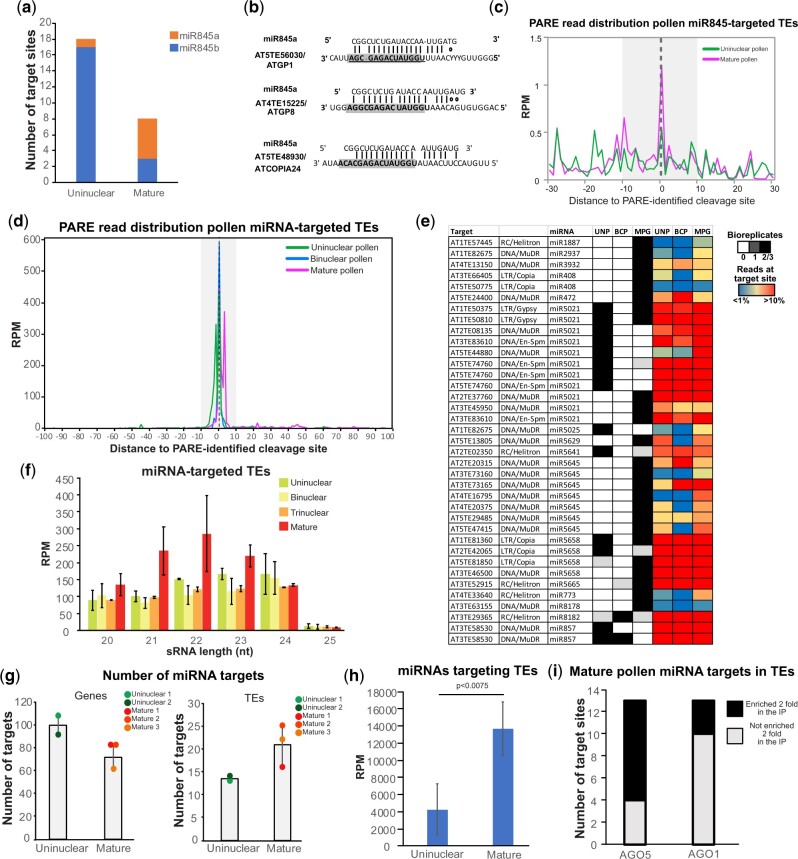
Figure 6.
Analysis of miRNA TE targets in the uninuclear, binuclear, and mature pollen grain identified by PARE sequencing. A, Number of miR845a- and miR845b-targeted TEs identified in uninuclear and mature pollen grain PARE libraries using high-confidence parameters. B, Representative examples of miR845a targeting positions in TEs. The location of the two different PBSs is highlighted in gray. C, Distribution of 5′-ends of PARE reads around the confirmed miR845-targeted TEs cleavage site (located at coordinate 0 in the X-axis) in a 30-nt window. Gray zone represents the physical position covered by the bound miRNA. D, Distribution of 5′-ends of PARE reads around the confirmed miRNA-targeted TEs cleavage site (located at coordinate 0 in the X-axis) in a 100-nt window. Gray zone represents the physical position covered by the bound miRNA. E, High-confidence identified miRNA-targeted TEs in PARE libraries. Target TE and its associated miRNA are indicated together with a heatmap of the presence of the target site in the different number of unicellular or mature PARE libraries in different shades of gray, and the level of accumulation of PARE reads in a 20-nt window surrounding the miRNA target site compared to the total accumulation of PARE reads for that gene for each individual bioreplicate studied with a blue-red gradient. F, Accumulation size profile of TE-derived siRNAs of PARE identified miRNA-targeted TEs. G, Number of targeted genes (left) and TEs (right) in uninuclear and mature pollen PARE libraries. Values for each bioreplicate are shown. H, Total accumulation values (in reads per million) of TE-targeting miRNA families (shown in (B) and including miR845) in uninuclear and mature pollen sRNA libraries. P-value was obtained using a paired t test. I, Number of identified TE-miRNA target sites in the mature pollen grain enriched more than two-fold (black) or not enriched (white) in AGO5 and AGO1.