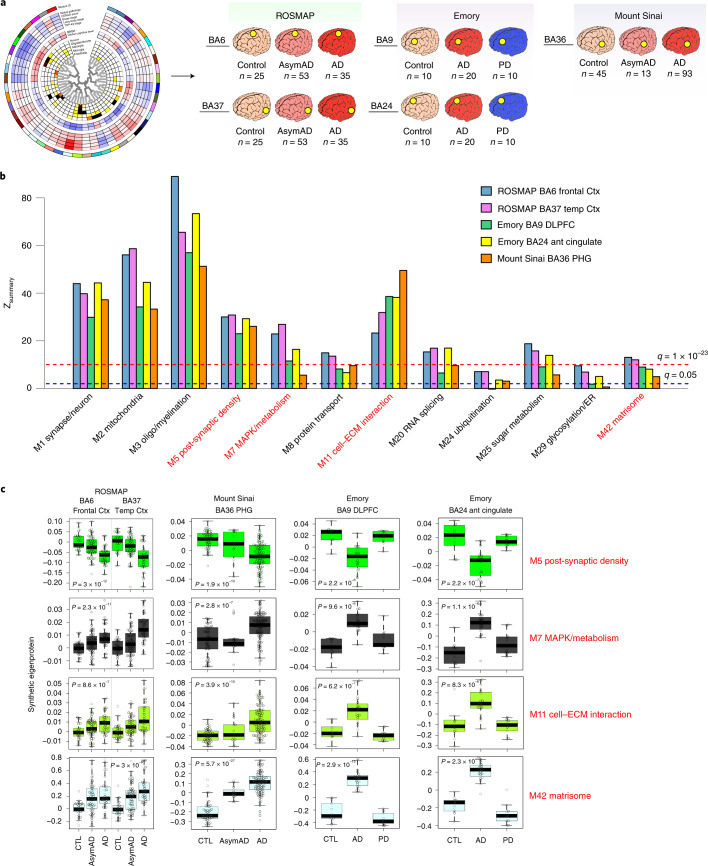
Fig. 2. Preservation of the TMT AD network across different cohorts, centers, methods and brain regions.
a–c, Module preservation and synthetic eigenprotein analysis of the TMT AD network generated from DLPFC BA9 tissues was performed in ROSMAP BA6 (frontal cortex) and BA37 (temporal cortex), Emory BA9 (DLPFC) and BA24 (anterior cingulate) and Mount Sinai Brain Bank BA36 PHG tissues (a). The Emory tissues included PD cases and were analyzed using a different TMT quantification approach (SPS-MS3). The Mount Sinai tissues were processed and analyzed by MS2-based TMT-MS at a different center. b, Module preservation of the 12 trait-correlated modules highlighted in Fig. 1b,c. Modules that had a Zsummary score greater than or equal to 1.96 (or q = 0.05, blue dotted line) were considered to be preserved, whereas modules that had a Zsummary score greater than or equal to 10 (or q = 1 × 10−23, red dotted line) were considered to be highly preserved. Preservation statistics for all TMT AD network modules are provided in Extended Data Fig. 3. c, Module eigenprotein level by case status was assessed in the different cohorts and brain regions shown in a by measuring a TMT AD network synthetic eigenprotein, representing the top 20% of module proteins by module kME, in each cohort and region. Synthetic eigenprotein levels are shown for the four most highly trait-correlated TMT AD network modules. Differences and statistics for all modules are provided in Supplementary Table 17. ROSMAP BA6 n = 25 control, 53 AsymAD, 35 AD; ROSMAP BA37 n = 25 control, 53 AsymAD, 35 AD; Emory BA9 n = 10 control, 20 AD, 10 PD; Emory BA24 n = 10 control, 20 AD, 10 PD; Mount Sinai BA36 n = 45 control, 13 AsymAD, 93 AD. Differences in synthetic eigenprotein levels were assessed by one-way ANOVA. Box plots represent the median and 25th and 75th percentiles, and box hinges represent the interquartile range of the two middle quartiles within a group. Data points up to 1.5 times the interquartile range from the box hinge define the extent of whiskers (error bars). ant, anterior; CTL, control; Ctx, cortex, temp, temporal.