Figure 3.
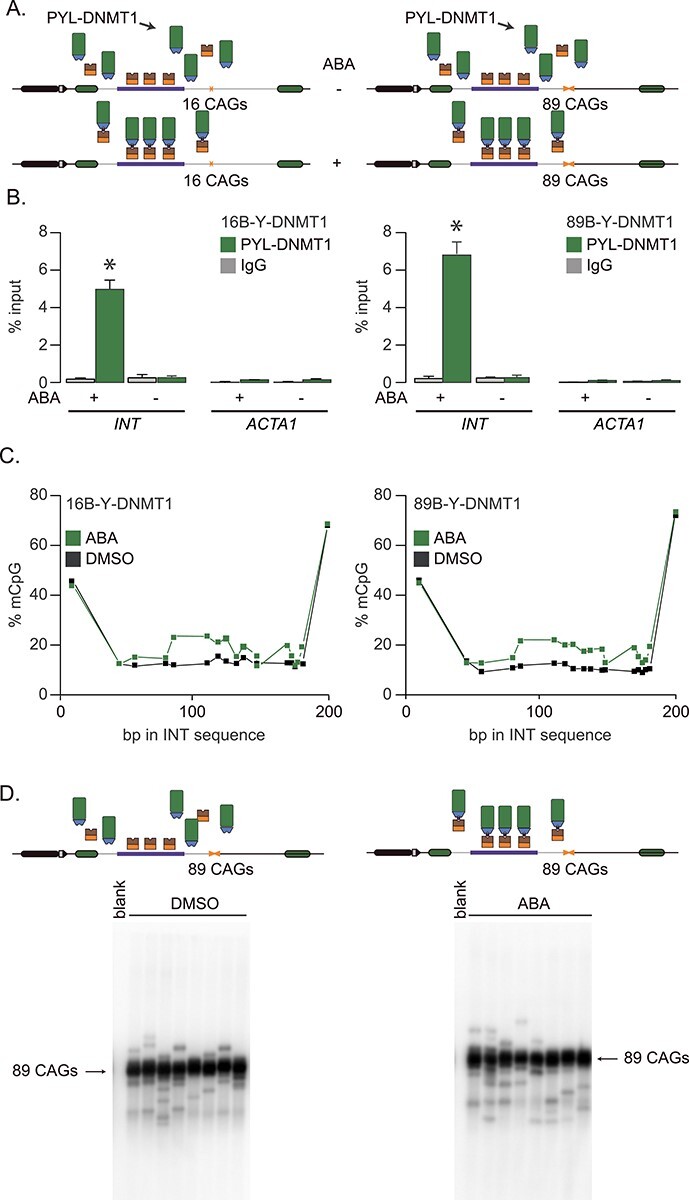
Inducible targeting of PYL-DNMT1 leads to changes in CpG methylation. (A) Schematic representation of 16B-Y-DNMT1 (left) and 89B-Y-DNMT1 (right) cell lines. (B) ChIP-qPCR using antibodies against FLAG to pull down PYL at INT and ACTA1 in 16B-Y-DNMT1 cells (left, N = 4) and 89B-Y-DNMT1 cells (right, N = 4).after 5-day treatments with ABA or DMSO. The error bars represent the standard error. (C) Bisulfite sequencing showing the percentage of methylated CpG motifs at the INT sequence in 16B-Y-DNMT1 (left) and 89B-Y-DNMT1 (right) cells in the presence of DMSO alone (black) or ABA (green) after 5-days of continuous ABA or DMSO treatment. (D) Representative SP-PCR blots after 30 days of continuous culture in the presence of DMSO (left) or ABA (right) in 89B-Y-DNMT1 cells. One nanogram of DNA/reaction used in both cases.
