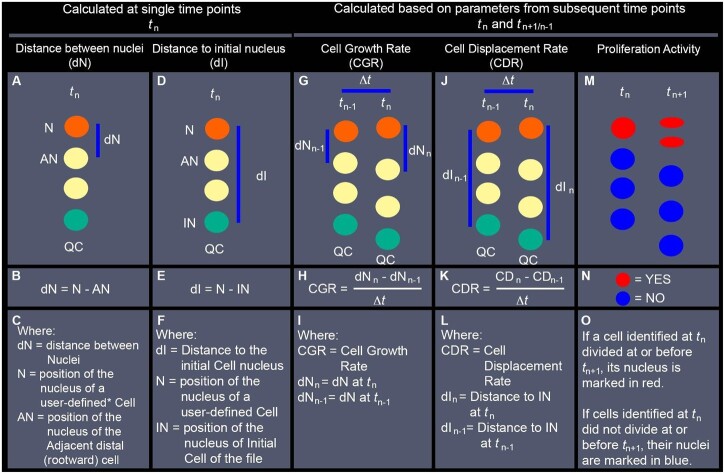
Figure 6.
Description of nuclear position-based parameters evaluated with LiPlaCeT plugin followed by analyses with ParaView. Simplified diagrams represent nuclei of cells within a single cell file. A–C, Estimation of the distance between adjacent nuclei within the same file (dN) at the displayed time point (tn). D–F, Distance to the nucleus of the initial cell of the file (dI), estimated as the distance between a user-defined Nucleus (N)* and the nucleus of the initial cell of the file (IN), at the displayed time point (tn). G–I, CGR, approximation calculated as the rate at which estimated dN changed during a period between the displayed time (tn) and a previous time point (tn–1) (The Methodology followed for evaluation of CGR in scenarios when cell division occurs between tn–1 and tn, is described in Supplemental Figure S1). J–L, CDR, approximation calculated as the rate at which each nucleus was displaced away from the IN during a period between the displayed time (tn) and a previous time point (tn−1). M–O, Distribution of cell proliferation events that took place between the displayed time point (tn) and the following timepoint (tn+1). QC position is indicated in all diagrams as a positional reference, but its position is not used in the calculations of the described parameters. Instead, the position of the nucleus of the initial cell (IN, green circles) is used as a reference for the distal limit of each cell file. * a user-defined cell (mentioned in (C)), is an individual cell identified manually using the LiPlaCeT plugin. Every nucleus manually marked acquires a unique ID containing its position and lineage information at different time points. This information is in turn analyzed and displayed for each user-defined nucleus using ParaView.