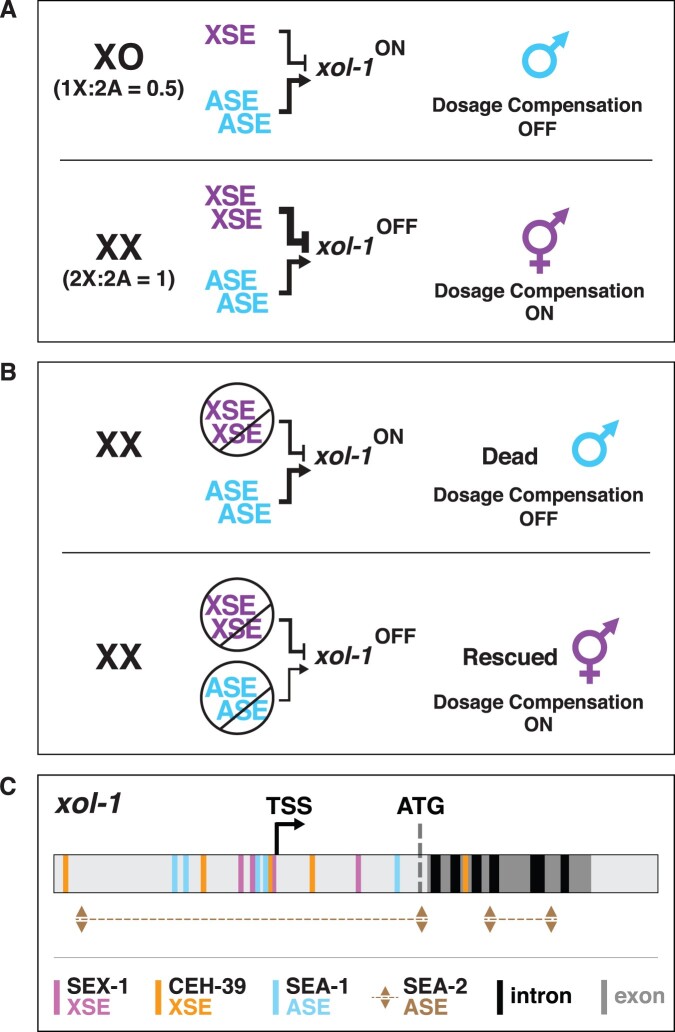
Figure 3.
Dissecting the X:A sex determination signal. (A) XSE regulate xol-1 in a dose-dependent manner in the context of two doses of ASE. Two doses of XSEs win out and repress xol-1 in diploid animals with two doses of ASE, which stimulate xol-1 expression. One XSE dose does not prevail in repressing. When xol-1 is activated in 1X:2A animals, the dosage compensation machinery is turned off. XO animals are viable and develop as males. When xol-1 is repressed in 2X:2A animals, the dosage compensation machinery is activated, thereby reducing X-linked gene expression by half. XX animals are viable and develop as hermaphrodites. (B) Loss-of-function mutations in XSEs were identified in genetic screens because they caused a xol-1 reporter transgene to be activated in XX animals, resulting in the masculinization and death of XX animals. XSEs were also discovered as suppressors of the male lethality caused by duplication of large regions of X. Loss-of-function mutations in ASEs were identified in genetic screens because they suppressed the lethality of mutations in XSEs and prevented the transformation of sexual fate caused by them. (C) Locations of binding sites in the 5′ xol-1 regulator regions for the XSEs (SEX-1 and CEH-39) that repress xol-1 transcription and the ASEs (SEA-1 and SEA-2) that activate xol-1 transcription. The general regions of SEA-2 binding were defined but not yet the precise binding sites. SEX-1 is a nuclear hormone receptor; CEH-39 is a ONECUT homeodomain protein; SEA-1 is a T-box protein; SEA-2 is a zinc-finger protein. After the molecular tug-of-war to control xol-1 transcription, a second tier of regulation occurs to control xol-1 pre-mRNA splicing by the XSE FOX-1 (see Figure 5).