Figure 1.
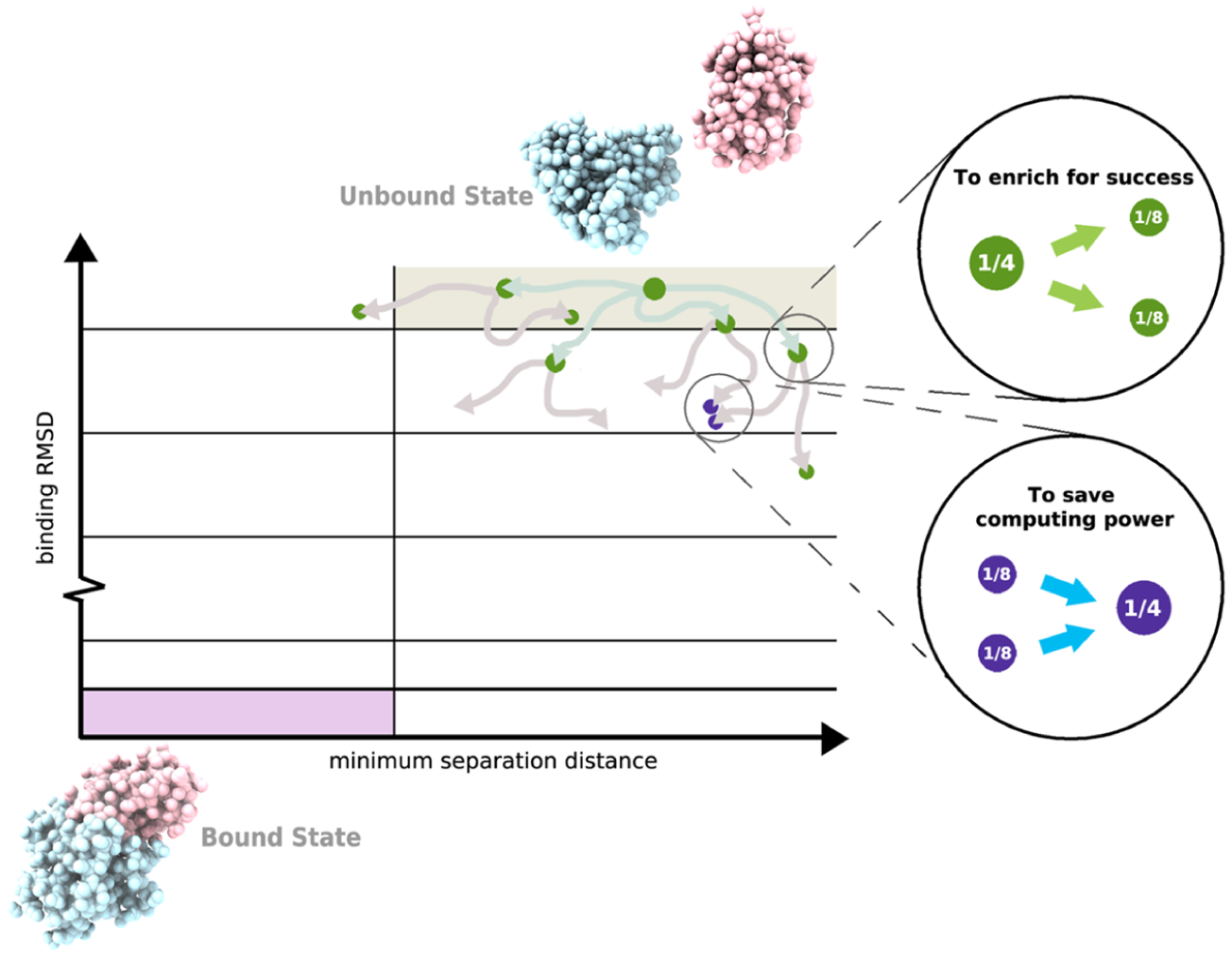
Basic WE protocol. As illustrated for the simulation of a protein–protein binding process, a two-dimensional progress coordinate is divided into bins with the goal of occupying each bin with a target number of four trajectories. Four equally weighted trajectories are initiated from the unbound state and subjected to a resampling procedure at periodic time intervals τ for the following: (i) to enrich for success, trajectories that make transitions to less-visited bins are replicated to generate a target of four trajectories in these bins, splitting the weights evenly among the child trajectories (green spheres) and (ii) to save computing time, the lowest-weight trajectories in bins that have exceeded four trajectories are terminated, merging their weights with those of higher-weight trajectories in these bins (purple spheres). Spheres are sized according to their statistical weights.
