Figure 6.
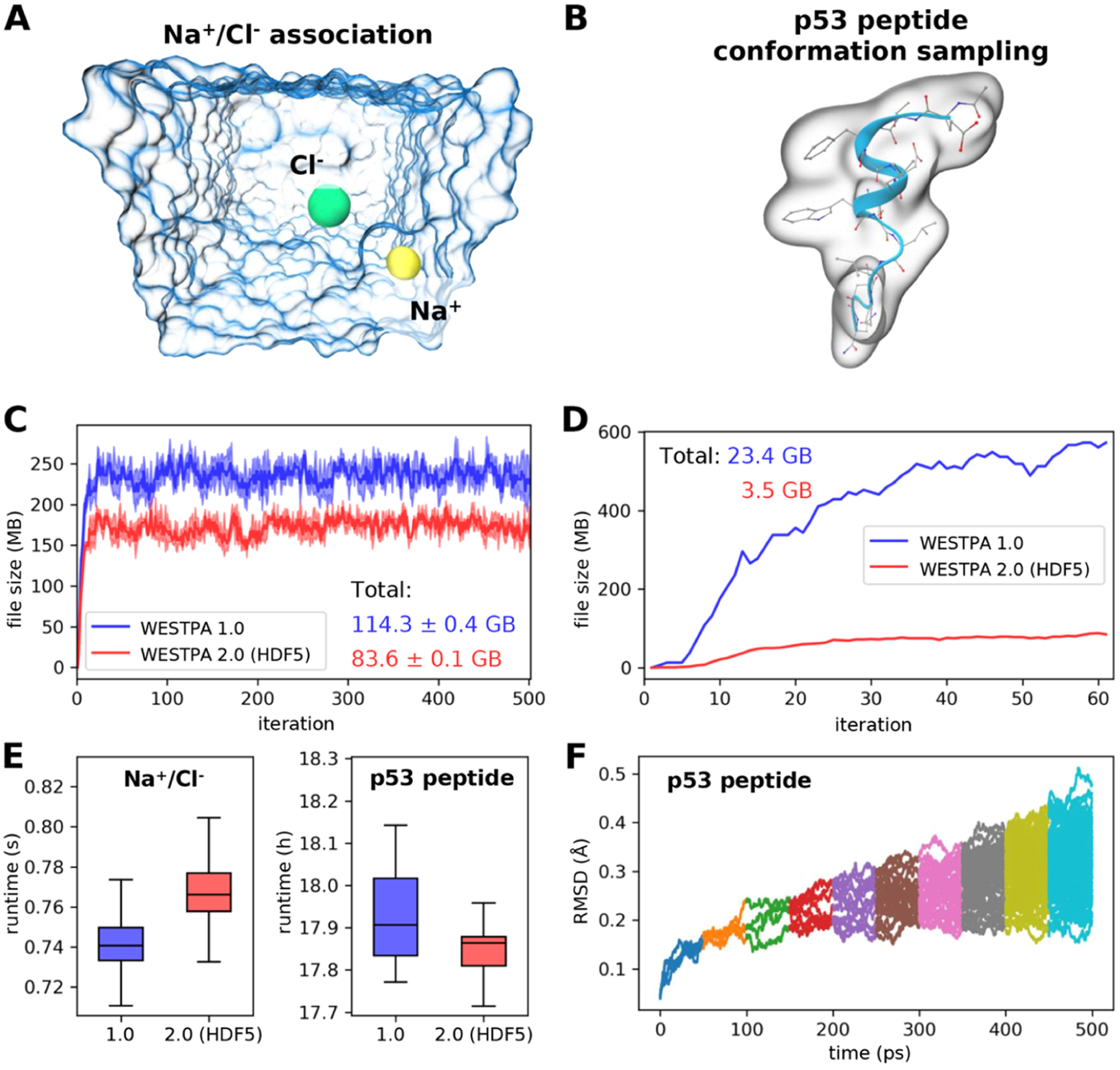
Demonstration of the usage of the HDF5 framework for two example systems. (A) Na+/Cl– association simulation where Na+ (yellow sphere) and Cl– (green sphere) ions were solvated in explicit water (blue transparent surface). The distance between the two ions serves as the progress coordinate. (B) Conformational sampling of a p53 peptide (residues 17–29) in a generalized Born implicit solvent using a progress coordinate consisting of the heavy-atom root mean square deviation (rmsd) of the peptide from its MDM2-bound conformation.21 The molecular surface of the p53 peptide is rendered as a transparent surface, with both the secondary (blue ribbon) and atomic structures overlaid. (C) Comparison of file sizes of per-iteration HDF5 files for the Na+/Cl– association simulation as a function of the WE iteration using WESTPA 1.0 and 2.0 with the HDF5 framework. The result was obtained from three independent simulations where the solid curves show the mean file sizes, while the light bands show the standard deviations. (D) Same comparison as panel (C) for a single simulation of the p53 peptide; hence, no error bars are shown. (E) Comparison of wall-clock runtimes normalized by the number of trajectory segments per WE iteration using WESTPA 1.0 and 2.0 with the HDF5 framework option turned on. (F) Time evolution of the heavy-atom rmsd of the p53 peptide from its MDM2-bound conformation using trajectories obtained using WESTPA’s analysis tools. Colors represent rmsds obtained from different iterations. WESTPA simulations of Na+/Cl– association and the p53 peptide were run using the OpenMM 7.5 MD engine.41
