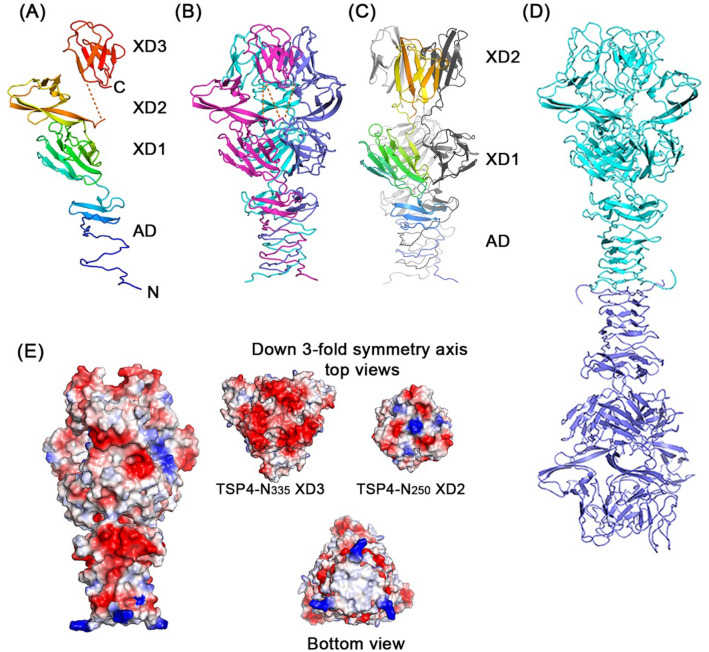
Figure 2.
Structures of TSP4-N335 and TSP4-N250. A cartoon representation of (A) TSP4-N335 monomer shown with spectrum color from blue N-terminus to red C-terminus. (B) TSP4-N335 trimer highlighting the three monomers in different colors. (C) TSP4-N250 trimer shown with one subunit in spectrum colors and two subunits in gray. The XD3 domain is missing. The spectrum color range span rainbow colors from blue to orange, to highlight the difference in the XD2 domain locations compared with that seen in TSP4-N335 as shown in (A). (D) Dimer of TSP4-N335 trimers. The two trimers associate via their N-terminal surfaces perpendicular to a shared threefold symmetry axis of the triple-stranded α-helix. (E) Left: Surface vacuum electrostatic potential calculated using PyMol with red color depicting negatively charged regions and blue color depicting positively charged regions. The trimer is viewed along the threefold symmetry axis. Right: Three views down the threefold symmetry axis. The top images show the XD3 surface on the left (based on the TSP4-N335 structure) and the XD2 surface on the right (based on the and TSP4-N250 structure that lacks XD3). The bottom image shows the N-terminal hydrophobic patch (white color) of the AD domain that mediates protein–protein interaction. Side chains that were not associated with electron densities during the refinement were omitted from the experimental structures. However, to fully account for all the charges and dipoles that contribute to the electrostatic potential, these side chains were added with favorable conformations.