Abstract
Macrophages (MΦs) display remarkable plasticity and the ability to activate diverse responses to a host of intracellular and external stimuli. Despite extensive characterization of M1 MΦs and a broad set of M2 MΦs, comprehensive characterization of functional phenotype and associated metabotype driving this diverse MΦ activation remains. Herein, we utilized an ex vivo model to produce six MΦ functional phenotypes. Isolated CD14+ PBMCs were differentiated into resting M0 MΦs, and then polarized into M1 (IFN-γ/LPS), M2a (IL-4/IL-13), M2b (IC/LPS), M2c (IL-10), and M2d (IL-6/LIF) MΦs. The MΦs were profiled using a bioanalyte matrix of four cell surface markers, ~50 secreted proteins, ~800 expressed myeloid genes, and ~450 identified metabolites relative to M0 MΦs. Signal protein and expressed gene profiles grouped the MΦs into inflammatory (M1 and M2b) and wound resolution (M2a, M2c, and M2d) phenotypes; however, each had a unique metabolic profile. While both M1 and M2b MΦs shared metabotype profiles consistent with an inflammatory signature; key differences were observed in the TCA cycle, FAO, and OXPHOS. Additionally, M2a, M2c, and M2d MΦs all profiled as tissue repair MΦs; however, metabotype differences were observed in multiple pathways including hexosamine, polyamine, and fatty acid metabolism. These metabolic and other key functional distinctions suggest phagocytic and proliferative functions for M2a MΦs, and angiogenesis and ECM assembly capabilities for M2b, M2c, and M2d MΦs. By integrating metabolomics into a systems analysis of MΦ phenotypes, we provide the most comprehensive map of MΦ diversity to date, along with the global metabolic shifts that coorelate to MΦ functional plasticity in these phenotypes.
Keywords: Macrophage, Immunomodulation, Metabolism, Plasticity
Summary Sentence
Characterization of ex vivo macrophage functional phenotypes by cell-surface marker expression, signal protein secretion, and transcriptomics correlate with distinct metabolic processes.
Graphical Abstract
INTRODUCTION
Macrophages (MΦs) display remarkable functional plasticity and the ability to activate diverse responses to a host of intracellular triggers, external stimuli, and nutritional availability. Generally, MΦs are divided into the well-defined the M1 (classically activated, pro-inflammatory) MΦs and a broad set of M2 (alternatively activated, anti-inflammatory) MΦs. M2 MΦs have been further subdivided into M2a, M2b, M2c, and M2d subtypes [1]. These M2 subtypes have been shown to exhibited numerous, often overlapping, effector functions [2], and have been found in diverse environments including wounds [3], tumors [4–6], developing fetuses [7], and systemic autoimmune diseases [8, 9]. Comprehensive, functional characterization of such a diverse array of MΦ populations presents an immense challenge and requires utilization of a systems-based approach. The emergent field of immunomodulation has established a pivotal role for metabolism in MΦ polarization and functional phenotype (reviewed in O-Neill, et al. [10]); yet the majority of this research is limited to the M1 and M2a phenotypes, leaving much to be learned about the other M2 MΦs.
Classically activated, pro-inflammatory M1 MΦs are polarized by exposure to IFN-γ, TNF, LPS, and/or other antimicrobial signals that trigger TLR ligation [11]. Considered essential for infection defense, the phagocytosis of apoptotic cells, and the promotion Type I immunity, M1 MΦs exhibit increased and sustained pro-inflammatory responses through the production of anti-microbial molecules and inflammatory mediators [12]. The alternately activated. M2 MΦs broadly exhibit tissue-reparative, pro-fibrotic, pro-angiogenic, and phagocytic functions [13–15]. Associated with Type 2 immunity, M2 MΦs are proposed to counteract the inflammatory microenvironment created by the M1 MΦ phenotype [13]. M2a MΦs are polarized by exposure to IL-4 plus IL-13 and have been functionally described to promote helper T cell activation, suppress inflammation, recruit tissue-repair populations, and promote angiogenesis [13, 16, 17]. M2b MΦs are polarized by exposure to LPS plus immune complexes (ICs) and have been shown to crosslink Fcγ receptors via ICs, promote Type 2 immunity, and secrete high levels of IL-10 [13, 18, 19]. M2c MΦs, polarized by exposure to IL-10, glucocorticoids, or TGF-β, are characterized by increased expression of MerTK, surface display of CD163, secretion of PGF, and activation of MMPs to promote tissue repair through efferocytosis, ECM remodeling, and angiogenesis [14, 20]. Tumor associated MΦs (TAMs, referred to herein as M2d MΦs) can be polarized through a variety of mechanisms including co-culture with cancer cells or ascites fluid, exposure to IL-6, LIF, and/or the purine nucleoside adenosine [15]. Associated with potent immunosuppressive functions and the promotion of angiogenesis [15, 21], M2d MΦs have been shown to secrete numerous proteolytic enzymes (e.g., MMP-2), growth factors (e.g.,VEGF), and anti-inflammatory mediators (e.g., TGF-β) [4, 6, 15, 22].
Metabolic immunomodulation has been associated with MΦ polarization into both the M1 and M2a functional phenotypes [10]. M1 MΦ polarization favors aerobic glycolysis and glycolytic flux into the PPP to facilitate oxidative burst and rapid biosynthesis [10]. The anti-microbial function of M1 MΦs is further enhanced by a decoupled anaplerotic TCA with breaks at citrate and succinate. These breaks facilitate the biosynthesis of fatty acids (FAs), prostaglandins, the anti-microbial metabolite itaconate, the generation of NO, and the HIF-mediated expression of pro-inflammatory cytokine [23–27]. In contrast, M2a MΦs utilize glycolysis to feed an intact TCA cycle coupled with FA oxidation (FAO) and active OXPHOS [28, 29], reducing inflammation through reduced lipid accumulation [30]. Additionally, M2a MΦs rely heavily on glutamine for production of N-glycans [26] and arginine flux through ornithine for collagen production [26, 31].
Much less is known regarding metabolic immunomodulation for the remaining M2 MΦ phenotypes (i.e., M2b, M2c, and M2d). In general, alternately activated M2 MΦs have been shown to have an intact TCA, increased OXPHOS, low cellular energy (e.g. high AMP/ATP ratios), and elevated catabolism of FAs [32]; however, M2a and M2c MΦs could be distinguished by glutamine consumption [26], elevated glycolysis in M2d MΦs is proposed to promoted angiogenesis and tumor metastasis [4], and M2b MΦs generate NO combined with decreased production of urea in a similar fashion as M1 MΦs [13]. Clearly, much remains to be learned about the metabotype of these distinct functional phenotypes.
Multiple MΦ functional phenotypes have been described in the literature, as described above and reviewed in Anders, et. al., 2019 [33], yet comprehensive characterization of this functional diversity in primary human cells remains to be established and the associated metabolism driving immunomodulation has not been fully described. With a systems biology approach, we comprehensively examined an ex vivo MΦ model that included a parent, resting phenotype (M0) and produced five polarized functional phenotypes (M1, M2a, M2b, M2c, and M2d). To our knowledge, this is the most comprehensive functional phenotyping of the MΦ subtypes reported to date. In addition, by integrating metabolomics into a systems analysis of these MΦ functional phenotypes, we provide the associated metabotypes. The findings presented herein lay the foundation for ongoing investigation into the relationship between metabolism and immunomodulation.
MATERIALS AND METHODS
Human Subjects, Monocyte Isolation, and MΦ Differentiation and Polarization
Written informed consent was obtained from healthy human subjects (ages 18–60 years old) enrolled in the study approved by the VA Puget Sound institutional review board (IRB). Whole blood (100 mL total) was obtained through venipuncture; 70 mL of blood was drawn into 60-mL syringe tubes pretreated with heparin (Fresenius Kabi USA, LLC; Lake Zurich, Illinois, USA) while 30 mL was collected into BD Vacutainer Serum tubes (Franklin Lakes, NJ, USA). The vacutainer serum tubes were incubated at room temperature for 30–60 minutes before centrifugation at 3500 rpm for 20 minutes and separated serum was collected and stored at −20° C. PBMCs were obtained via Ficoll-paque density centrifugation (Ficoll-Paque Plus, GE Healthcare Bio-Sciences; Uppsala, Sweden) followed by CD14+ monocyte isolation via negative immunomagnetic selection using the Pan Monocyte Isolation Kit (Miltenyi Biotech; Bergisch Gladbach, Germany). Monocyte culture media was made of Roswell Park Memorial Institute (RPMI) 1640 (Corning; Manassa, VA, USA) modified with 1% (v/v) each of minimum essential media (MEM) non-essential amino acids (Gibco; Grand Island, NY, USA), sodium pyruvate (Gibco; Grand Island, NY, USA), L-glutamine (Lonza; Walkersville, MD, USA) and supplemented with 10% (v/v) autologous human serum (AHS; pooled from a minimum of four healthy human donors) and 50 ng/mL of recombinant human MΦ colony-stimulating factor (M-CSF) (Biolegend; San Diego, CA, USA). Monocytes were cultured in monocyte culture media at a cellular density of 1.5 × 106 cells/mL in 6-well cell culture plates. and incubated at 37° C with 5% CO2. Monocytes received fresh modified culture media on day 4 and day 7 and were treated with predefined polarization stimuli to achieve the desired MΦ phenotype. Cells exposed to M-CSF alone produce the M0 phenotype. Classically activated M1 phenotype was achieved by exposure to 100 ng/mL LPS from Escherichia coli O111:B4 (Sigma Aldrich; St. Louis, MO, USA) and 20 ng/mL recombinant human IFN-γ (PeproTech; Rock Hill, NJ, USA). Alternately M2 MΦs were activated as follows: M2a MΦs from stimulation with 20 ng/mL recombinant human IL −4 (PeproTech; Rock Hill, NJ, USA) and 25 ng/mL recombinant human IL-13 (PeproTech; Rock Hill, NJ, USA); M2b cells with 100 ng/mL LPS and 16.7 μL/mL immune complexes (IC; see details below); M2c cells with 25 ng/mL recombinant human IL-10 (PeproTech; Rock Hill, NJ, USA); and M2d MΦs from stimulation with and 50 ng/mL recombinant human IL-6 (R&D Systems; Minneapolis, MN, USA) and 25 ng/mL recombinant human leukemia inhibitory factor (LIF; R&D Systems; Minneapolis, MN, USA). ICs for M2b polarization were generated by mixing 1 μL of a 1.32 mg/mL solution of bovine serum albumin (BSA; MP Biomedicals; Auckland, New Zealand) with 50 μL anti-albumin (bovine serum), rabbit IgG fraction polyclonal antibodies (Invitrogen; Eugene, OR, USA) and incubating at 37° C for one hour. This IC mixture, stable at 4° C for several days, was stored until needed. Polarized cells were incubated at 37° C with 5% CO2 from between 24 and 72 hours in accordance with established MΦ differentiation protocols.
MΦ Functional Phenotyping
Flow Cytometry
Flow cytometry was conducted to access differences in cell surface marker expression between the six different phenotypes. Preliminary experiments were conducted using CD14+ monocytes on 2-, 5- and 7-days post-plating to differentiate between monocytes, immature MΦs, and mature M0 MΦs and to identify gating regions for future MΦ phenotype experiments. For MΦ phenotype experiments, a panel of four fluorescent markers was employed including fluorescein isothiocyanate (FITC) mouse anti-human CD40 (BD Biosciences; San Jose, CA, USA), phycoerythrin (PE) mouse anti-human CD86 (BD Biosciences; San Jose, CA, USA), peridinin chlorophyll protein complex cyanine (PerCP-Cy™5.5) mouse anti-human CD163 (BD Biosciences; San Jose, CA, USA) and allophycocyanin (APC) mouse anti-human human leukocyte antigen – DR isotype (HLA-DR) (BD Biosciences; San Jose, CA, USA). Unstained cells, single stained cells, and fluorescent minus one (FMO) controls were included for each analyzed phenotype to identify unstained cellular populations. All samples were evaluated on an Accuri C6 flow cytometer (BD Biosciences; San Jose, CA, USA) and the data analyzed using Kaluza 2.1 Analysis software (Beckman Coulter; Indianapolis, Indiana, USA).
Secreted Cytokines, Chemokines, and Growth Factors Detection
All cytokines, chemokines and growth factors were measured using the supernatant collected during the intracellular metabolite extraction at 24- and 72-hr post polarization. The analytes were evaluated utilizing multiplex magnetic bead-based immunoassays (ProcartaPlex™, Thermo Fisher Scientific; Waltham, MA, USA) per the manufacturer’s instructions, and were run on a MAGPIX® instrument (Luminex Corporation; Austin, TX, USA).
RNA Extraction
RNA was purified utilizing the Qiagen RNAeasy® plus mini kit (Germantown, MD, USA). The lysing tubes containing the interphase layer from the metabolite were first thawed, if needed, on ice, resuspended in the RLT lysis buffer supplemented with 1% (v/v) of β-mercaptoethanol and homogenized, as described above. Following lysis, the RNA was extracted and purified according to the manufacturer’s instructions. The integrity of the extracted RNA was evaluated on an Agilent 2200 Tape Station (Santa Clara, CA, USA) and diluted to a final volume of 5 μl, at a concentration of 20 ng/μL.
Gene Expression Profiling
The nCounter Gene Expression – Hs Myeloid v2 Panel CodeSet (NanoString Technologies; Seattle, WA, USA) was used to evaluate the expression of genes associated with myeloid innate immune system response. In brief, and according to NanoString’s procedure, 100ng of each RNA sample was added to the CodeSet in hybridization buffer and incubated at 65°C for 16 hours. Purification and binding of the hybridized complexes were then carried out automatically on the nCounter Prep Station using magnetic beads derivatized with short nucleic acid sequences that are complementary to the capture and reporter probes. The hybridization mixture was first allowed to bind to the magnetic beads via the capture probe then washed to remove excess reporter probes and unhybridized DNA fragments. Probe complexes were then eluted off the beads and hybridized to magnetic beads complementary to the reporter probe followed by wash steps to remove excess capture probes. In the last step, the purified target-probe complexes were eluted off and immobilized in the cartridge for data collection which was carried out in the MAX System Digital Analyzer (NanoString Technologies; Seattle, WA, USA). Digital images were processed, and barcode counts tabulated in a comma-separated value (CSV) format. The raw count data was then normalized to the mean of positive control probes followed by RNA content normalization to the geometric mean of housekeeping genes in the CodeSet. Data normalization was performed using nCounter Analysis Software version 4.0 (NanoString Technologies; Seattle, WA, USA). Gene set enrichment analysis (GSEA) was utilized to determine which Nanostring-defined functional pathways were regulated within the individually polarized phenotypes. Two representations of these pathways are presented in the data. The first, undirected global significance (UGS) score, was utilized to identify which pathways were differentially regulated in each phenotype regardless of whether the genes were up- or down-regulated when compared to the M0 MΦs. An extension of this score, the directed global significance statistics (DGS), measured the extent to which the genes were up- or down-regulated within the given pathway. In general, a positive pathway score for a specific phenotype indicates a predominance of genes that are up-regulated within the pathway as compared to those that are down-regulated; a negative pathway score indicates the reverse [34]. Additional analyses were conducted using bioDBnet Biological Database [35] and DAVID bioinformatics resources [36]. The top up-regulated genes within each activated phenotype were selected for gene ontology (GO) enrichment analysis. The GO processes significantly enriched (p < 0.05) were then chosen for comparison. With further evaluation, processes common to all phenotypes and related to either specific diseases or ubiquitous to the immune systems were eliminated from the GO process list (e.g. GO:0045824: negative regulation of innate immune response).
MΦ Metabotyping
Intracellular Metabolite Isolation
With the goal of creating a comprehensive snapshot of cellular function, multiple experimental sample types were collected from each MΦ phenotype during cell harvest to obtain transcriptomic, proteomic, and metabolomic data from each experimental trial. At the desired time point, the cellular supernatant, composed of cell culture media and any non-adherent cells, was removed from the cell culture well and placed into falcon tubes on ice. One mL of ice-cold phosphate buffered saline (PBS) was added to the adherent cell layers and then removed to the tubes containing the cellular supernatant and the tubes centrifuged at 2000 rpm for 8 minutes. The collected supernatant was stored at −80° C until further analysis. Pelleted cells were flash-frozen in liquid nitrogen and stored on ice, while the remaining adherent cells were quenched with 350 μL of ice-cold 100% methanol (Honeywell; Muskegon, MI, USA) and 350 μL of ice-cold ultrapure distilled water (Invitrogen; Grand Island NY, USA) and removed through gentle cell scraping and added to the cell pellet fraction. Following the addition of 700 μL of ice-cold chloroform (Acros Organics; Thermo Fisher Scientific; Waltham, MA, USA), the collected cells were vortexed for 30 seconds and were transferred to FastPrep® lysing matrix D tubes (MP Biomedicals; Auckland, New Zealand). To achieve cell lysis, the tubes were homogenized during two cycles of 40 s each at 4.0 m/s with a 90 s delay between cycles utilizing the FastPrep-24™ 5G Homogenizer (MP Biomedicals; Auckland, New Zealand). The homogenized samples were centrifuged at 16,000 x g for 5 minutes at 4° C, and then placed immediately on ice. The polar (methanol/water) layer and non-polar (chloroform) layers were subsequently transferred to 1.5 mL protein low binding microcentrifuge tubes. These metabolite suspensions were lyophilized overnight without heat on a Thermo Scientific™ Savant™ ISS110 SpeedVac™ (Waltham, MA, USA) and stored at −80°C until the samples were shipped to Metabolon for further analysis. The remaining interphase layer was flash-frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Metabolite Detection, Identification, and Quantification
All samples were analyzed by Metabolon (Morrisville, NC, USA) using four ultra-high-performance liquid chromatography/tandem accurate mass spectrometry (UHPLC/MS/MS) methods. The following is a summary of Metabolon’s procedure. All methods utilized a Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. Lyophilized samples were reconstituted in solvents compatible to each of the four methods and contained a series of standards at fixed concentrations to ensure injection and chromatographic consistency. One aliquot was analyzed using acidic positive ion conditions, chromatographically optimized for more hydrophilic compounds. In this method, the extract was gradient eluted from a C18 column (Waters UPLC BEH C18–2.1×100 mm, 1.7 μm) using water and methanol, containing 0.05% perfluoropentanoic acid (PFPA) and 0.1% formic acid. A second aliquot was also analyzed using acidic positive ion conditions that was chromatographically optimized for more hydrophobic compounds. In this method, the extract was gradient eluted from the same aforementioned C18 column using methanol, acetonitrile, water, 0.05% PFPA and 0.01% formic acid and was operated at an overall higher organic content. A third aliquot was analyzed using basic negative ion optimized conditions using a separate dedicated C18 column. The basic extracts were gradient eluted from the column using methanol and water, however with 6.5 mM ammonium bicarbonate at pH 8. The fourth and final aliquot was analyzed via negative ionization following elution from a HILIC column (Waters UPLC BEH Amide 2.1×150 mm, 1.7 μm) using a gradient consisting of water and acetonitrile with 10mM ammonium formate, pH 10.8. The MS analysis alternated between MS and data-dependent MS scans using dynamic exclusion. The scan range varied slighted between methods but covered 70–1000 m/z. Several types of controls were analyzed in concert with the experimental samples: a pooled matrix sample generated by taking a small volume of each experimental sample served as a technical replicate throughout the data set; extracted water samples served as process blanks; and a cocktail of quality control (QC) standards that were carefully chosen not to interfere with the measurement of endogenous compounds were spiked into every analyzed sample, allowed instrument performance monitoring and aided chromatographic alignment. Instrument variability was determined by calculating the median relative standard deviation (RSD) for the standards that were added to each sample prior to injection into the mass spectrometers. Overall process variability was determined by calculating the median RSD for all endogenous metabolites (i.e., non-instrument standards) present in 100% of the pooled matrix samples. Experimental samples were randomized across the platform run with QC samples spaced evenly among the injections.
Raw data was extracted, peak-identified and QC processed using Metabolon’s hardware and software. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities. Biochemical identifications are based on three criteria: retention index within a narrow RI window of the proposed identification, accurate mass match to the library +/− 10 ppm, and the MS/MS forward and reverse scores between the experimental data and authentic standards. The MS/MS scores are based on a comparison of the ions present in the experimental spectrum to the ions present in the library spectrum. Peaks were quantified using area-under-the-curve. The resulting data was then normalized to cell count to account for differences in metabolite levels due to differences in the amount of material present in each sample.
Metabolomics Data Analysis
After normalization, peak intensity values were uploaded into MetaboAnalyst 4.0 (Ste. Anne de Bellevue, Quebec) [37], normalized using pareto scaling, and then statistically analyzed. Using MetaboAnalyst 4.0, partial least-squares to latent structures discriminant analysis (PLSDA) and orthogonal PLSDA (OPLSDA) were employed to characterize and visual metabolomic differences between the resting MO MΦs and the individually polarized phenotypes. For detailed descriptions of OPLSDA utilization with metabolomics data, readers are referred to Wiklund, et al., (2008) [38]. PLSDA, a regression method, was employed to identify relationships between the UHPLC/MS/MS data (X) and binary vectors (Y) with a value of 0 for M0 resting MΦs and 1 representing the polarized phenotype as they are compared individually to the M0 cells. Orthogonal PLS (OPLS) was then used to separate the systematic variation in X into two components: the first component, a predictive component (covariance), linearly associated with Y, describes between phenotypic variance; and the second component (correlation), orthogonal to Y describes between sample variation [38].
To adequately demonstrate the OPLSDA results, three data visualization methods were utilized. The first, cross-validated score plots, display the cross-validated score values (Orthogonal T-score) on the y-axis versus the predictive or model T-score (T-score) on the x-axis for each sample point within the analysis. S-Plots were used to visualize the predictive component of the model. The x-axis represents the magnitude or covariance of each metabolite to the model while the reliability or correlation of those metabolites is plotted on the y-axis. Generally, metabolites that have covariance scores greater than 0.2 or less than −0.2. and correlation values greater than 0.6 or less than −0.6 are considered to contribute significantly to the predictive value of the model. Shared and Unique Structure (SUS) plots were employed to compare the results from the M1 OPLS model to each M2 (M2a, M2b, M2c and, M2d) phenotype model. For example, by plotting the correlation from the predictive component from the M0 versus M1 model and the M0 versus M2a model, metabolic biomarkers can be identified that are not only uniquely correlated to the M1 and M2a MΦs, but also have shared differentiation from the parent M0 phenotype. The correlation scores for each M2 subtype were plotted on the y-axis and the M1 scores on the x-axis. In this representation, metabolite features that are shared between phenotypes will cluster close to the diagonals while those features unique to each phenotype will fall outside of the diagonals.
Metabolomics Data Deposition
Global metabolomics data was deposited in the National Metabolomics Data Repository accessed through the Metabolomics Workbench (metabolomicsworkbench.org) and supported by the NIH Common Fund Metabolomics Program. Original scale metabolite value by raw area count and associated metadata are deposited under Study ID ST001835 and will be released from embargo upon publication.
Online Supplemental Material
Additional data provided in the Online Supplemental Material includes gene and protein expression of immunomodulatory factors (Supplemental Figure 1). Supplemental Figure 2 details global transcriptomics differences between each MΦ functional phenotype and the parent M0 phenotype. Supplemental Figure 3 includes representations of PCA and OSPLDA multivariate analysis of the global transcriptomic data and a heatmap demonstrating metabolic differences between each MΦ functional phenotypes relative to the parent, resting macrophage phenotype. Additional phenotypic data for the for γ-glutamyl amino acids and various lipid subgroups are depicted in Supplemental Figure S4. Supplemental Tables S1 and S2 outline the relative genetic expression of select transcripts and the relative concentrations of select pathway metabolites for each polarized phenotype compared to M0 parent cells, respectively.
RESULTS
Classification of MΦ Phenotypes by Cell Surface Markers Provides Poor Resolution of Subtypes
Six MΦ phenotypic subpopulations were derived from CD14+ monocytes isolated from human PBMCs and differentiated into resting MΦs (referred to herein as M0 MΦs) prior to polarization into M1 (IFN-γ/LPS), M2a (IL-4/IL-13), M2b (IC/LPS), M2c (IL-10), and M2d (IL-6/LIF) MΦ phenotypes. For simplicity of notation, these phenotypes will be referenced herein as M1, M2a, M2b, M2c, and M2d MΦs. Following polarization, the parent non-polarized MΦ population and five polarized MΦ subpopulations were functionally phenotyped using a systems biology approach.
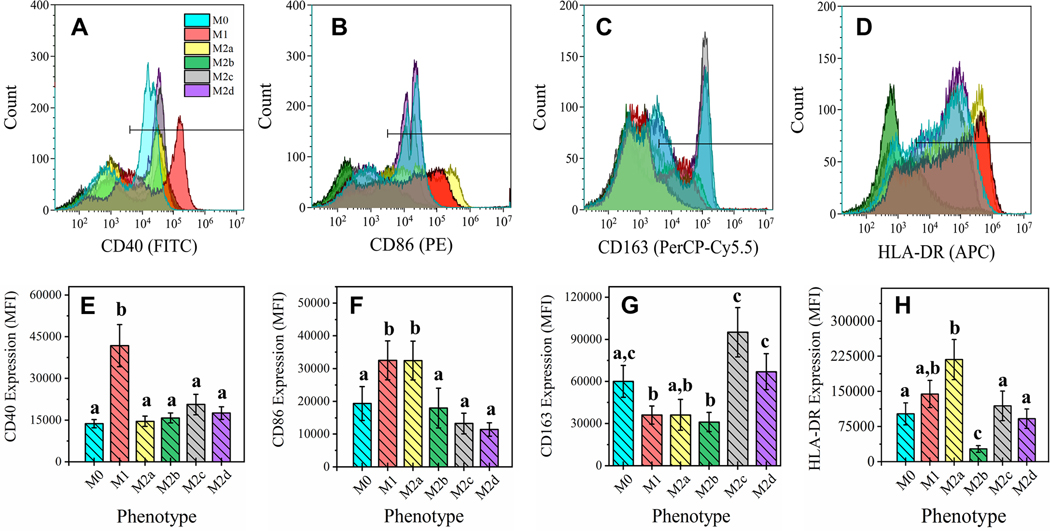
Four MΦ cell surface markers known to distinguish functionality [39] were used as a first-pass characterization of phenotype (CD40, CD86, CD163, and HLA-DR). As shown in the histogram overlay (Fig. 1 A-D) and associated MFI bar charts (Fig. 1E-H), pro-inflammatory M1 MΦs (red bars) demonstrated significantly higher cell-surface display for CD40 (p < 0.05; Fig 1A, E) and CD86 (p < 0.05; Fig. 1B, F) relative to resting MΦs (M0, blue bars). Within the alternatively activated MΦ subgroups, the M2a MΦs (yellow bars) displayed significantly higher levels of both CD86 (p < 0.05; Fig. 1B, F) and HLA-DR (p < 0.05; Fig. 1D, H) upon polarization from the M0 MΦs, indicating APC functionality. Cell-surface display of CD163 (p < 0.05; Fig. 1C, G) was elevated in M2c (gray bars) and M2d MΦs (purple bars), while all other phenotypes had decreased cell-surface display, specifically to a significant level in M1 and M2b MΦs, relative to M0 MΦs. Interestingly, M2b MΦs (green bars) had a significantly decreased cell-surface display of HLA-DR to all other MΦ phenotypes, not just relative to the M0 phenotype (p < 0.05; Fig. 1D, H). Taken together, M1 MΦs profiled as CD40highCD86highCD163low, M2a MΦs as CD86highCD163lowHLA-DRhigh, M2b MΦs as CD163lowHLA-DRlow, while both M2c and M2d MΦs appeared as CD86lowCD163high.
Figure 1: Macrophage Functional Phenotyping by Common Cell-Surface Markers.
Cell surface marker profile of six MΦ phenotypes were detected by FACS. CD14+ monocytes were isolated from human blood-derived PBMCs, differentiated into resting MΦ (M0, shown in blue) with M-CSF ex vivo, and polarized into five activated phenotypes using IFN-γ/LPS (M1, shown in red), IL-4/IL-13 (M2a, shown in yellow), IC/LPS (M2b, shown in green), IL-10 (M2c, shown in gray), or IL-6/LIF (M2d, shown in purple) for 72 hours. FACS gating on four common MΦ cell surface markers for all six MΦ phenotypes is shown as live cell count for each marker gate (A-D) and mean fluorescence intensity (MFI) profile of gate (E-H). The cell surface markers depicted include CD40 (A, E), CD86 (B, F), CD163 (C, G), and HLA-DR MHC class II receptor (D, H). Statistical analysis was performed for the MFI values (histogram bars) using repeated measures analysis of variance and model-based means post hoc test (p < 0.05) with differing letters denoting statistical significance. Histograms are total cell count and representative of three biological replicates. MFI is normalized to total cell count and includes all three biological replicates (N=3, mean ± SEM).
Targeted Profiling of Immune-Mediator Profile by both Transcript and Protein Expression Demonstrates MΦ Functional Phenotype Along Pro-/Anti-Inflammatory Axis
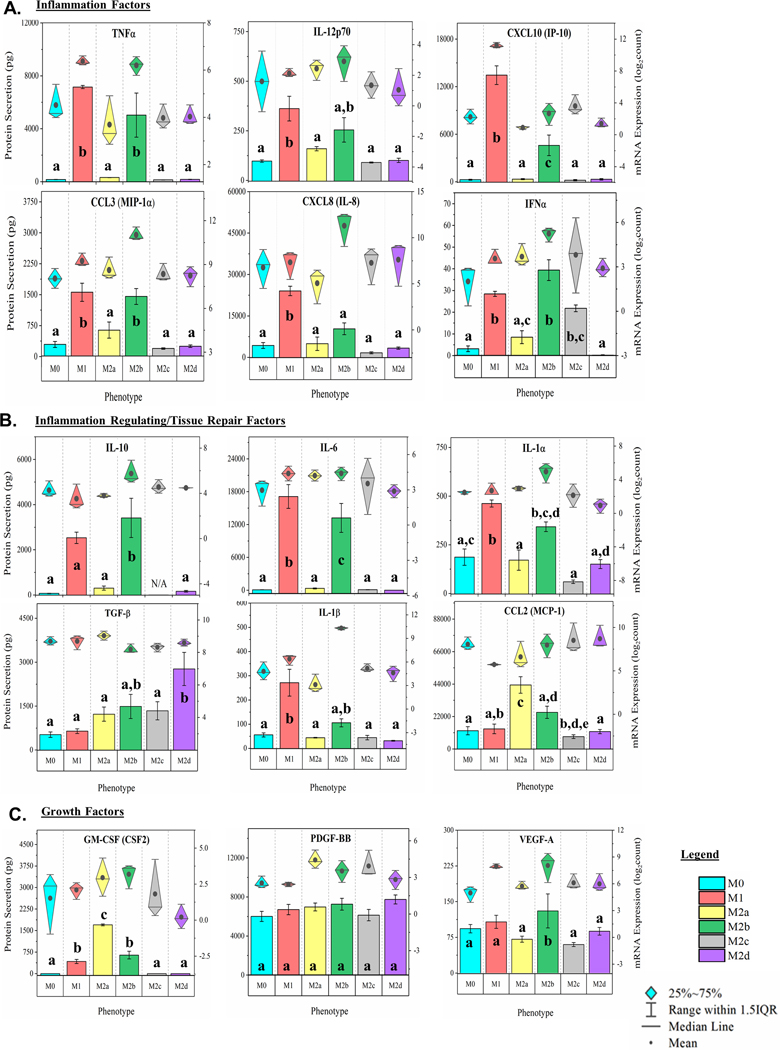
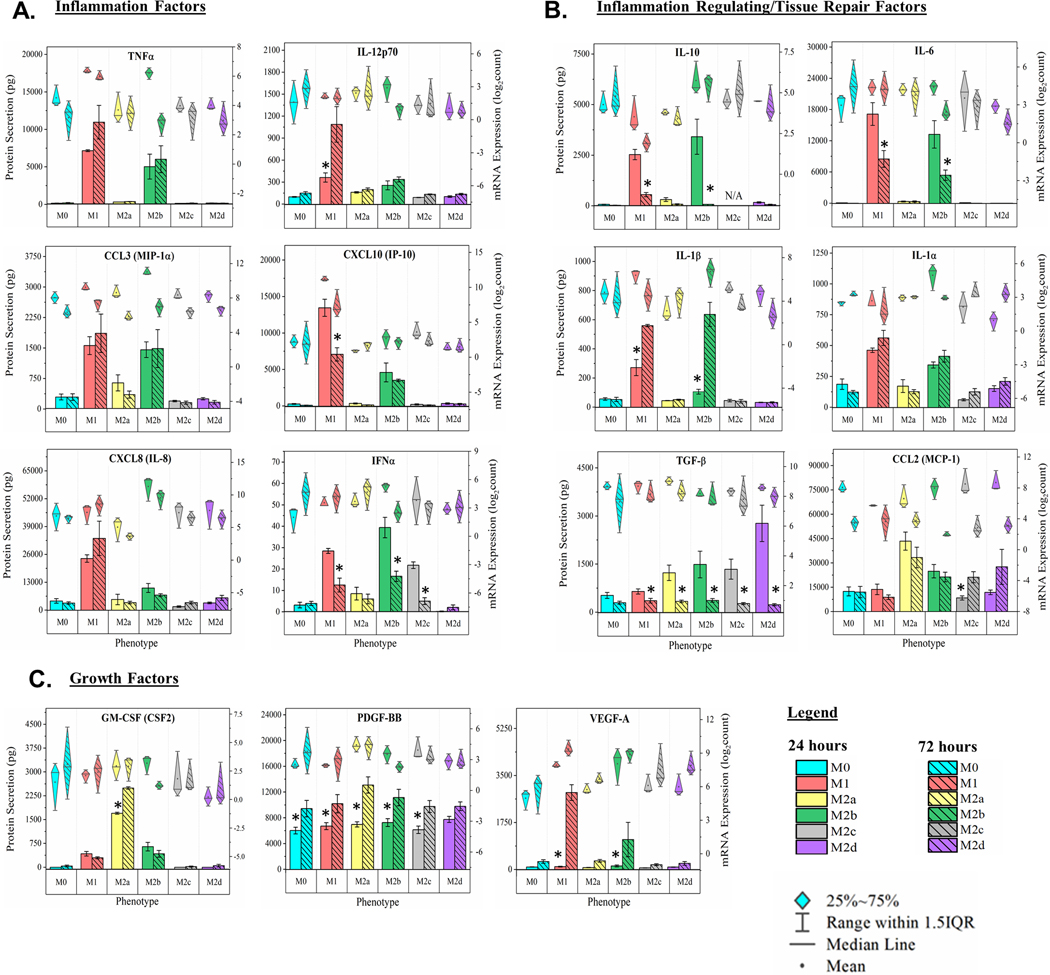
Cytokines, chemokines, and growth factors induced during MΦ polarization are involved in multiple effector functions, including the promotion or inhibition of inflammation. Post-polarization, MΦ culture supernatant was multiplex profiled for 22 immunomodulating proteins as depicted (Fig. 2A-C, bar charts, left y-axis and Supplemental Fig. 1A-G in the Online Supplementary Material). In parallel, total RNA was extracted from each MΦ culture and profiled for mRNA expression (Fig. 2A-C, diamond-whisker plots, right y-axis and Supplemental Fig. 1A-G). As IL-10, IL-4, IL-13, IFNγ, and IL-6 were used as polarizing factors for M2c, M2a, M1, and M2d MΦs, respectively, an N/A was used for these specific phenotypes and respective protein concentration profiles.
Figure 2: MΦ Functional Phenotyping by Gene and Protein Expression of Immunomodulatory Factors.
Multiplex detection of immunomodulatory factors in six MΦ phenotypes harvested at 24 hours were detected using magnetic bead-based quantification of mRNA and secreted protein. CD14+ monocytes were isolated from human blood-derived PBMCs, differentiated into resting MΦ (M0, shown in blue) with M-CSF ex vivo, and polarized into five activated phenotypes using IFN-γ/LPS (M1, shown in red), IL-4/IL-13 (M2a, shown in yellow), IC/LPS (M2b, shown in green), IL-10 (M2c, shown in gray), or IL-6/LIF (M2d, shown in purple). Gene and protein expression profile (diamond-whisker plots and bar charts, respectively) of key functional molecules were profiled by multiplex assay after 24 hours of ex vivo polarization. Immunomodulatory function includes pro-inflammatory (TNF⍺, IL-12p70, CCL3, CXCL10, CXCL8, and IFN⍺) (A), immune-regulatory/tissue-repair (IL-10, IL-6, IL-1b, IL-1⍺, TGF-b, and CCL2) (B), and growth factors (GM-CSF, PDGF-BB, and VEGF-A) (C). Diamond-whisker plots display 25%−75% quartile range, median, and mean. Bar charts indicate mean ± SEM. Statistical analysis was performed for the MFI values (histogram bars) using repeated measures analysis of variance and model-based means post hoc test (p < 0.05) with differing letters denoting statistical significance. Expression profiles were normalized to total cell count and include three biological replicates (N=3).
Of the pro-inflammatory cytokine and chemokine factors (p < 0.05; Fig 2A, Supplemental Fig. 1B), both M1 MΦ and interestly, the M2b MΦs secreted TNFα, CXCL10 (IP-10), CCL3 (MIP-1α), IFNα, and CCL4 (MIP-1β) at levels significantly higher than the M0 phenotype upon polarization. Relative to each other, M1 MΦs were distinguished from M2b MΦs by producing signifantly higher levels of CXCL10 (IP-10) and CXCL8 (IL-8). Of the other M2 MΦ subtypes, only the M2c phenotype secreted IFNα at statistically significant higher levels relative to the M0 MΦs. For cytokines and chemokines associated with tissue repair and anti-inflammatory properties (p < 0.05; Fig. 2B; Supplemental Fig. 1D, E) M1 and M2b MΦs secreted significantly higher levels of IL-6, IL-1α, IL-1β, IL-4, and IL-13 relative to M0 MΦs. M2a and M2d MΦs were distinguished from M0 MΦs by significantly higher levels of secreted CCL2 (MCP-1) and TGF-β, respectively.
Growth factors also play a key role in the wound healing process; however, only M2b MΦs were observed to secrete significantly higher levels of VEGF-A relative to the M0 MΦs (p < 0.05; Fig. 2C). M1, M2a, and M2b MΦs were observed to secrete statistically significant levels of GM-CSF (CSF2), relative to the M0 MΦs, with M2a MΦs secreting significantly more GM-CSF then either the polarized M1 or M2b MΦs (p < 0.05; Fig. 2C). While PDGF-BB secretion by tissue MΦs is hypothesized to promote tissue remodeling [40], significant differences in secretion of this growth factor were not observed with polarization (Fig. 2C).
Finally, the mean mRNA expression (represented by the dark circles in the diamond-whisker plots) generally reflect trends observed in secreted protein levels with a few notable exceptions. It is important to note that the detection of protein is on a linear scale (left x-axis), while gene expression is on a log scale (right x-axis), thus minimizing the appearance of differences in gene expression. Regardless, dramatic differeneces between gene expression and protein secretion are obserserved. For example, IL-1β and CXCL8 (IL-8) transcripts in M2b MΦs are significantly upregulated when compared to the other phenotypes, but secreted protein levels are significantly lower than the M1 MΦ profile (p < 0.05; Fig. 2 A, B). These striking differences support the proposed importance of post-transcriptional regulation to phenotypic functionality [41], specifically in the M2b MΦ phenotype.
Global patterns of myeloid gene expression demonstrate impact of polarization and further define functional phenotype as distinct from the parent, resting MΦ phenotype
Univariant, hierarchical clustering (HC) of gene expression levels revealed the modulatory effect polarization has on these MΦ phenotypes. The most impactful dynamics in gene expression were observed in M1, M2a, and M2b MΦs (Supplemental Fig. 2A). Interestingly, M1 and M2b clustered together, while all other M2 subtypes clustered more closely with their non-polarized parent phenotype M0 with M2a and M2c MΦs forming a unique cluster apart from M2d and M0 MΦs. In general, the relative number of differentially expressed genes (−1≤ Log2FC ≥1; Supplemental Fig. 2B) observed for each phenotype mirrors the patterns identified through HC, with M1 and M2b MΦs most transcriptionally active and M2a, M2c, and M2d decreasingly active in that order. The increased transcriptional activity observed in both M1 and M2b MΦs is primarily accounted for in upregulated gene expression while a fairly evenly distribution between up and down regulated genes was detected in the other M2 MΦ subtypes.
When the HC analysis was constrained to SDEGs via ANOVA analysis (p-value or false discovery rate [FDR]) of 0.05), additional cluster separation was demonstrated for the M2c and M2d phenotypes apart from the M2a and parent M0 MΦs (Supplemental Fig. 2C). Multivariate clustering by PCA supported these findings and demostrtated that the M1 and M2b MΦ phenotypes share a single dimension and are distinctly different from the other MΦ phenotypes further emphasizing the similar impact of polarization on gene expression patterns in these two phenotypes (Supplemental Fig. 3A).
The supervised OPLSDA provided clearer class separation of the polarized MΦ phenotypes from the M0 parent phenotype evidenced by clear phenotype cluster separation (x-axis) in the cross-validated score plots (Supplemental Fig. 3B-F). In agreement with the SDEG constrained HC and PCA analyses, between phenotype variance is greatest for M1 MΦs and M2b MΦs, relative to M0 MΦs, accounting for ~40–45% of the total variance (Supplemental Fig. 3B, D), and nearly twice the percent variance for M2a, M2c, and M2d MΦs at ~20–30% variance relative to M0 MΦs. Taken together, these data further demonstrate that when looking beyond the classic pro-/anti-inflammatory axis for defining MΦ phenotype, clear distinctions can be made that suggest phenotype plasticity includes a wide range of functionality in these cell subtypes.
Pathway Analysis and GO Process Characterization Indicates Biological Functionality Within Each Distinct MΦ Phenotype
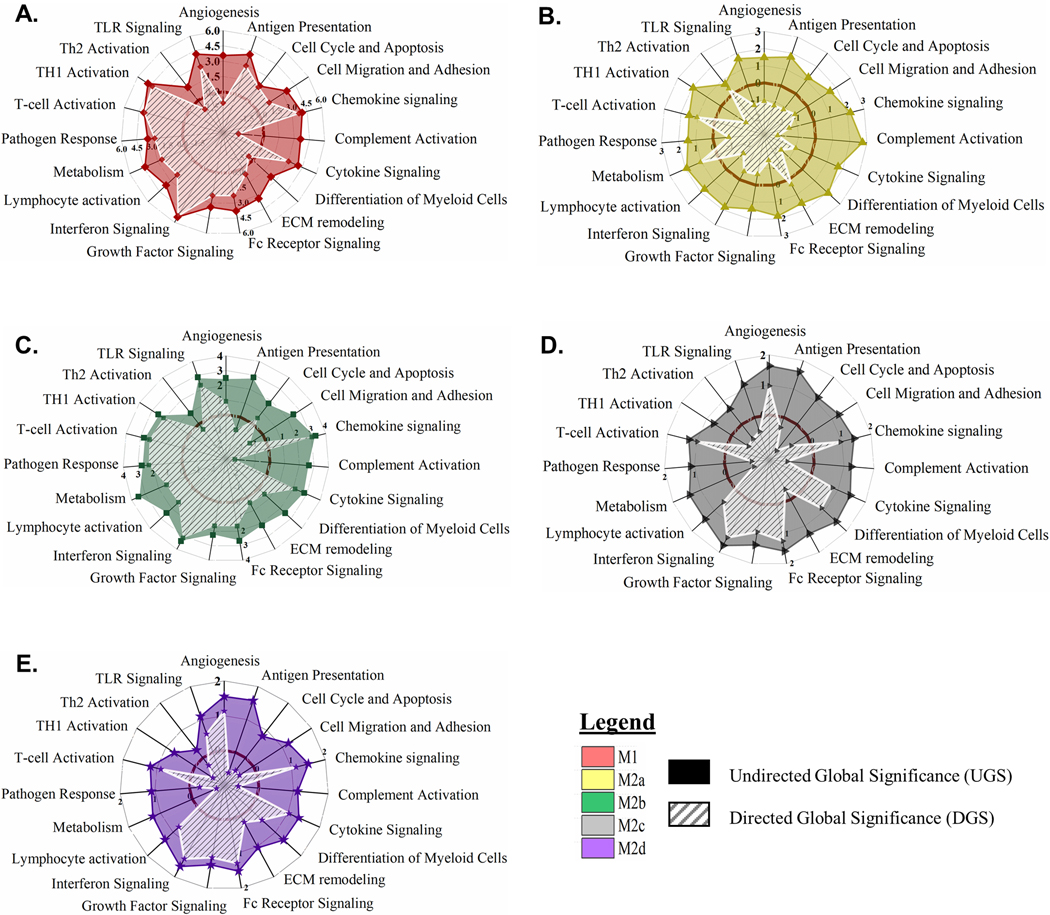
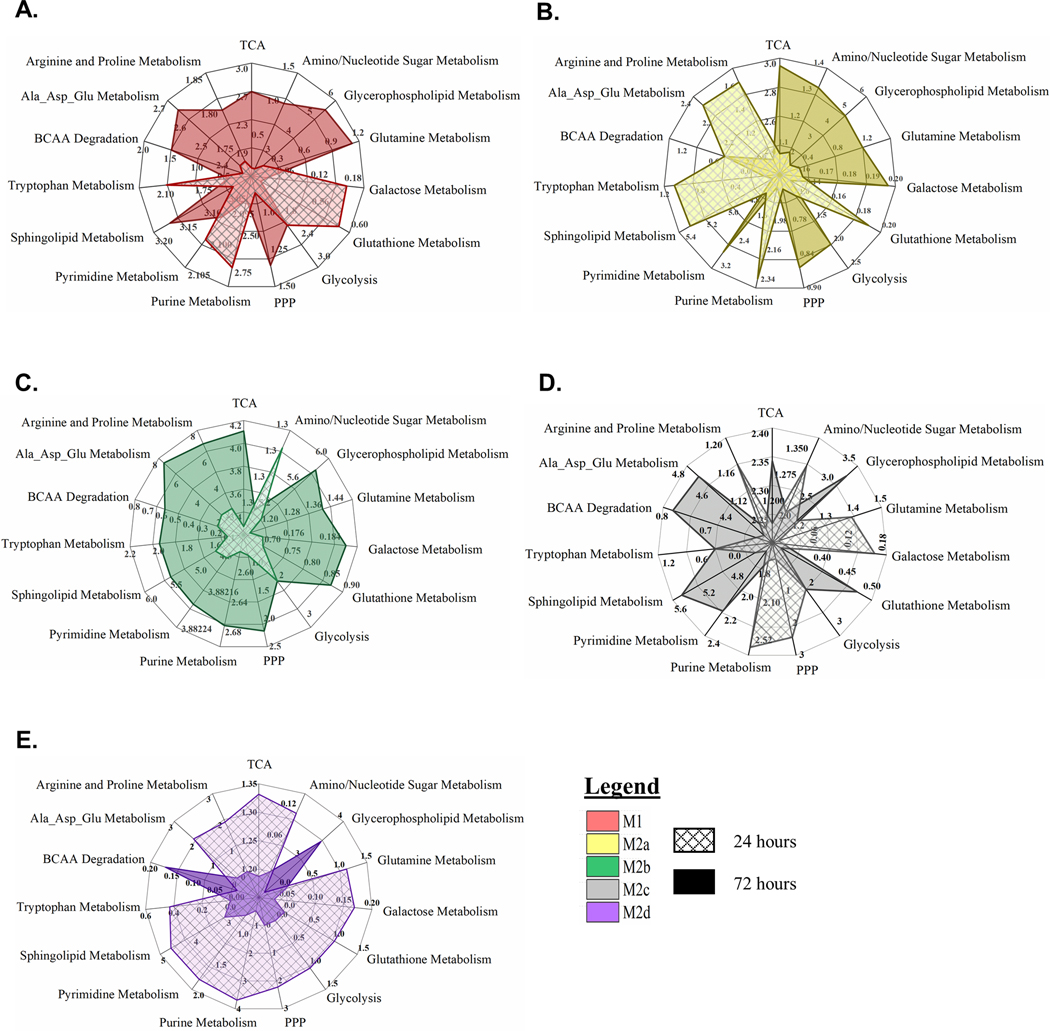
Gene set enrichment analysis (GSEA) was utilized to determine how transcriptional activation translated into putative myeloid functionality separate from the M0 MΦ phenotype. Undirected global significance scores (UGS) revealed 19 functionally defined, biological pathways differntially impacted relative to the resting M0 MΦs (displayed as solid colored plots in the radar graphs in Fig. 3). Consistent with the HC results, M1 and M2b MΦ polarization most dramatically impacted these functional pathways (Fig. 3A, C) with less significant impacts observed for the M2a, M2c, and M2d MΦs (Fig. 3B, D, E). The directed global significance statistic (DGS; displayed as white hashed plots in the radar graphs in Fig. 3), illustrated that polarization is an “activation” process for M1, M2b, and M2d MΦs with gene expression driving 15 biological pathways into activation for M1 MΦs and M2b MΦs (Fig. 3 A, C) and 11 pathways for M2d MΦs (Fig. 3E). In contrast, M2a MΦ and M2c MΦ gene expression pushed fifteen and ten biological pathways into “inhibition”, respectively (Fig. 3B, D). Highlighted SDEGs of relevance to these biological pathways are depicted in Supplemental Table 1.
Figure 3: MΦ Polarization into Functional Phenotypes Activates Distinct Gene Expression Profiles Relative to the Parent, Resting MΦ Phenotype (M0).
Gene set enrichment analysis (GSEA) and pathway impact scoring of global myeloid gene expression demonstrates distinct patterns in five polarized MΦ phenotypes. Briefly, multiplexed myeloid gene expression was directly detected through molecular barcode probes and normalized to the geometric mean of the housekeeping gene set. GSEA for each phenotype was calculated for pathway impact relative to the non-polarized, resting MΦ (M0). Radar plots for each polarized MΦ phenotype display pathway score based on impact regardless of whether genes were up- or down-regulated (Undirected Global Significance [UGS], solid color) and pathway score based on impact incorporating t-statistic comparison of gene regulation as relatively increased or decreased (Directed Global Significance [DGS], hatch marks). The dark maroon circle marks the “zero” impact score to differentiate activation versus inhibition of the designated pathways. Radar plots include UGS and DGS scores for all five polarized MΦ phenotypes: M1 MΦs (IFN-γ/LPS treated, shown in red) (A), M2a MΦs (IL-4/IL-13 treated, shown in yellow) (B), M2b MΦs (IC/LPS treated, shown in green) (C), M2c MΦs (IL-10 treated, shown in gray) (D), and M2d MΦs (IL-6/LIF treated, shown in purple) (E). GSEA based on three experimental replicates for each MΦ s phenotype (N=3).
M1 MΦ polarization resulted in gene expression patterns that drove functionality in anticipated directions with up-regulation of antigen presentation, cell migration and adhesion, chemokine signaling, cytokine signaling, interferon signaling, pathogen response, and Th1-specific T-cell activation pathways, while angiogenesis, complement activation, and Th2-specific pathways were downregulated (Fig. 3A). GSEA identified 97 SDEGs ((−1≤ Log2FC ≥1 and p < 0.05) contributing to 41% of IFN-γ responsive and 12% LPS-responsive gene expression. Within the IFN-γ responsive genes, interferon pathway genes, IRF1, IRF7, IRF8 and ISG15, are known transcriptional enhancers of genes associated with antigen presentation and Th1 responses (e.g. CD40 and CD86), cytokine and chemokine signaling (e.g. TNF, CXCL9, CXCL11, CCL4, CCL8, and STAT1), and pathogen response (e.g. IL18 and IRG1) [42]. These SDEGs and related upregulated pathways are consistent with defined pro-inflammatory and antimicrobial effector functions known to be associated with M1 MΦ polarization.
The IL-4 and IL-13 stimuli utilized for M2a polarization are generally considered mild polarization activators [43]. Consequently, M2a MΦs exhibited a remarkably different pathway profile than the M1 MΦs. While all 19 pathways were differentially regulated, only the T-cell activation, Th2-specific T-cell activation, metabolism, and ECM remodeling pathways were upregulated (Fig. 3B). Of the 83 identified SDEGs, only 36% of these transcripts were upregulated. The most upregulated genes, CCL17, CCl22 and CCL18, support the generally accepted role that M2a MΦs play in cellular proliferation, ECM remodeling, and tissue repair. Additionally, known ECM remodeling metalloproteinase genes, ADAM19 and MMP12, and the common M2a MΦ-associated metabolism transcripts of TGM2, FABP4, and PTGS1 were also upregulated; however, the upregulated T-cell and Th2 activation pathways did not exhibit any SDEGs using our inclusion criteria.
As with our cluster analysis, M2b MΦ proposed biological functionality is unique from all other M2 MΦ subtypes and most like the M1 MΦ phenotype. In contrast to M1 MΦs, M2b MΦ gene expression during polarization resulted in the downregulation of antigen presentation and cell migration and adhesion response pathways (Fig. 3C). Chemokine signaling, the most upregulated pathway, is associated with prominent SDEGs and is intertwined with many other functional pathways. For example, chemokines CXCL2, CXCL3, and CXCL5 promote angiogenesis through the recruitment of angiogenic neutrophils and CCL5, CCL8, and CXCL9 support the recruitment and differentiation of myeloid cells necessary for tissue remodeling functions. While M2b MΦ polarization indicates biological functionality comparable to M1 MΦs, distinct differences in activated pathways indicate that polarization of this phenotype may have a more localized biological relevance.
In a similar trend to M2a MΦs, the pathway scores observed for M2c MΦs imply that IL-10 is also a mild polarizing stimulus; however, patterns of pathway up- and down-regulation are distinctly different. Of the 19 biological pathways detected, nine were differentially upregulated compared to the M0 MΦs including the upregulation of angiogenesis, chemokine signaling, cytokine signaling, differentiation of myeloid cells, Fc receptor signaling, growth factor signaling, interferon signaling, lymphocyte activation, and T-cell activation (Fig. 3D). Interestingly, many of the identified SDEGs have broad effects that regulate inflammation (DUSP1, SOCS3, S100A8, and S100A9), vascular insult and neutrophil chemotaxis (S100A8 and S100A9), as well as protection against apoptosis and oxidative stress (IER3) [14]. Based on these pathway impact profiles, the biological functionality of polarization into this MΦ phenotype may serve to modulate immune response in the transition to resolution.
At first glance, M2c and M2d MΦs appeared to polarize into phenotypes of similar biological functionality; however, distinct differences between these phenotypes were present (Fig. 3D, E). While their transcriptional profiles suggested overlapping effector functions such as resolution of inflammation (DUSP1, SOCS3, THBD, and CD163) and immune suppression (CCL8), the upregulation of TLR signaling in M2d MΦs during polarization was notably distinct from polarization in M2c MΦs. Additionally, M2d MΦs differentially regulated many known pro-angiogenic markers such as ENPP2, SPHK1, CXCL2, CXCL3, and ID1, findings in line with a proposed biological role beneficial to survival of associated tumors.
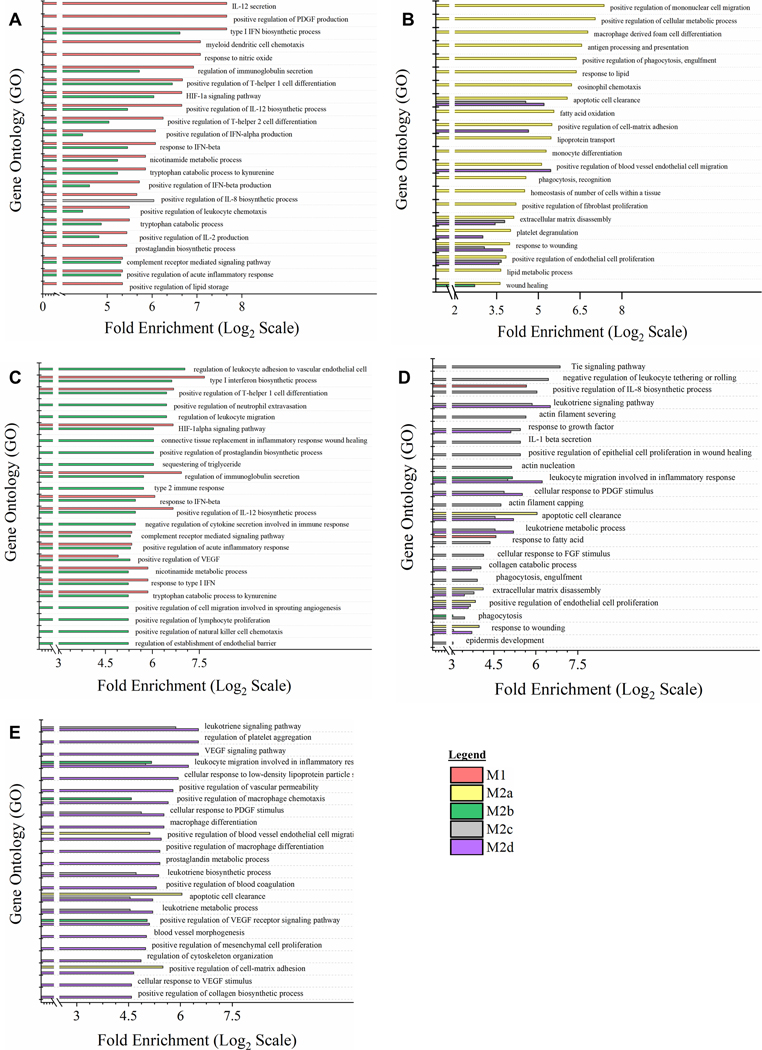
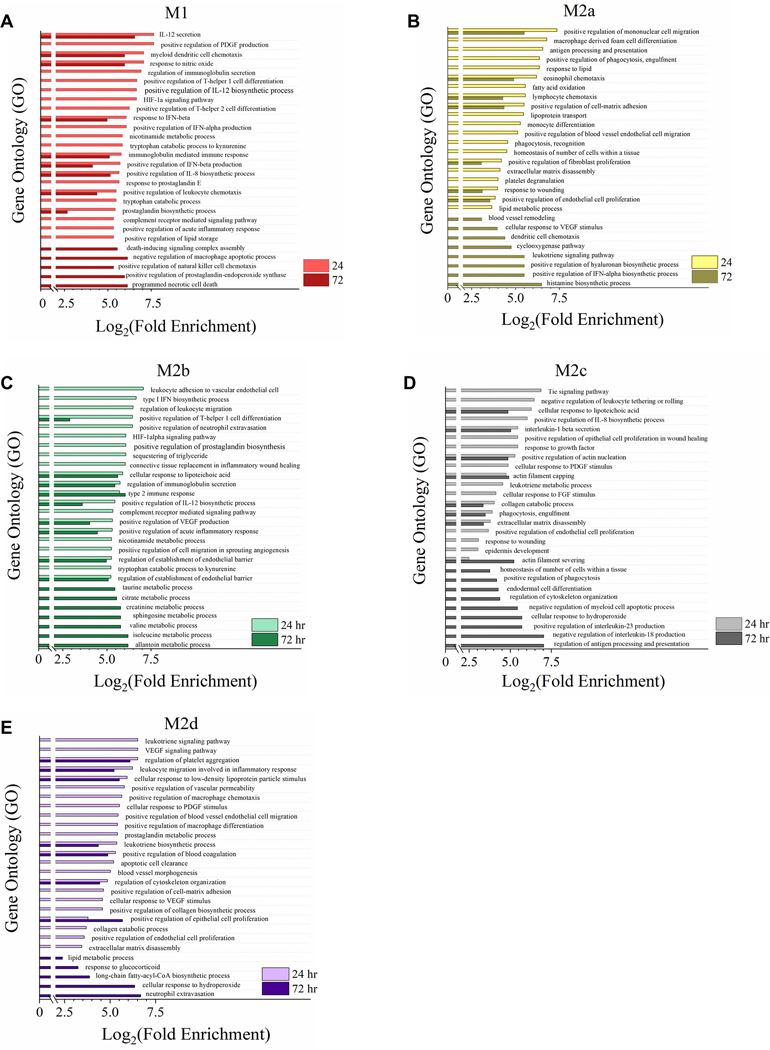
While UGS and DGS scores provided both magnitude and directionality of polarization of each MΦ phenotype overlaid on proposed biological functionality, GO annotation of global myeloid gene expression was utilized to provide a direct comparison between each polarized phenotype (Fig. 4). For each GO annotation, all five MΦ phenotypes are displayed; however, the immediate similarity between M1 and M2b MΦs can be observed when the GO annotation is selected for the top 15 GO processes for M1 MΦs and M2b MΦs. GO annotation generally supported DGS analysis of polarization; however, interesting differences were noted. Specifically, secretion of IL-12, regulation of PDGF production, and response to NO were pathways identified as unique to M1 MΦs (Fig. 4A), while regulation of leukocyte migration, neutrophil extravasation, and connective tissue replacement in inflammatory wound healing were pathways identified as unique to M2b MΦ (Fig. 4C).
Figure 4: Gene Expression Profile Comparison of Polarized MΦ Phenotypes Identifies Unique and Overlapping Immunomodulatory Functions.
Gene Ontology (GO) annotation of global myeloid gene expression in five MΦ phenotypes identifies relative biological functionality across polarized states. Multiplexed myeloid gene expression profile of all five polarized MΦ phenotypes was used to annotate impacted biological functionality based on GO Processes. For each MΦ phenotype, the top 15 impacted GO processes significantly enriched (p < 0.05) were plotted relative to the other four polarized MΦ phenotypes. Enrichment plots for prioritized GO process profile include all five polarized MΦ phenotypes: M1 MΦs (IFN-γ/LPS treated, shown in red) (A), M2a MΦs (IL-4/IL-13 treated, shown in yellow) (B), M2b MΦs (IC/LPS treated, shown in green) (C), M2c MΦs (IL-10 treated, shown in gray) (D), and M2d MΦs (IL-6/LIF treated, shown in purple) (E). GO process annotation was based on three experimental replicates for each MΦ phenotype (N=3).
Finally, of the other three M2 MΦ subtypes, GO processes associated with M2a MΦs (Fig. 4B) heavily favored tissue-reparative, pro-fibrotic, pro-angiogenic, and phagocytic functions, while the upregulation of scavengers CD163 and MerTk (Supplemental Table 1) for M2c MΦs are consistent with phagocytosis GO processes detected. Other M2c MΦ identified GO processes included the Tie signaling pathway, known to play a key role in angiogenesis (Fig. 4D). Similarly, for M2d MΦs nine of the top 17 GO processes were causally related to angiogenesis. Of the remaining eight listed processes, six contributed to ECM remodeling (Fig. 4E). In summary, both patterns of polarization detected through GSEA and DGS (Fig. 3) and direct comparison between phenotypes by GO annotation (Fig. 4) demonstrate unique and overlapping proposed biological functionality and further support the reorganization of M2b MΦs from being a M2 subtype to being more closely related to M1 MΦs in functionality.
Global Metabolomics Demonstrates Functional MΦ Phenotypes are also Distinct Metabotypes
Global metabolomics profile of the parent, resting M0 MΦs and five polarized MΦ phenotypes identified 498 compounds of known identity. The top 50 metabolites, as determined by ANOVA analysis (p < 0.05) and depicted in the heatmap (Supplemental Fig. 3G) demonstrated the immunomodulation of the individual phenotypes. The most striking distinction in the HC analysis is the metabolic differences, especially within lipid metabolism, observed between the M1 MΦs, the M0 MΦs and the M2 subtypes. Of specific interest is the clustering of the M2b MΦs with the other M2 subtypes considering the M2b MΦ propensity towards the M1 pro-inflammatory signature. The notable exception to this trend is the apparent similarities in M1 and M2b MΦ nicotinamide and tryptophan metabolism common to pro-inflammatory responses. Additionally, MΦ metabotype clustering was demonstrated through OPLSDA and represented in the cross-validated score plots (inset plots in Fig. 5A.1 - E.1). Separation along the x-axis (between phenotype variation) in the cross-validated score plots quantifies metabotype cluster separation between each polarized MΦ phenotype and the parent M0 MΦs, with cluster separation greatest for M2b MΦs (14.2%) and M1 (13.0%) and the least for M2d (10.3%) and M2c (9.4%) and M2a (8.1%) MΦs. The y-axis scores (orthogonal T score or within phenotype variation) reflected the notable variation observed within each MΦ phenotype; however, the variation was similar for each phenotype (~60%). Furthermore, the larger within phenotype variance observed in this data warrants additional analysis and may indicate uncertainty in the metabotype prediction on the basis of cross-validation scores alone [38].
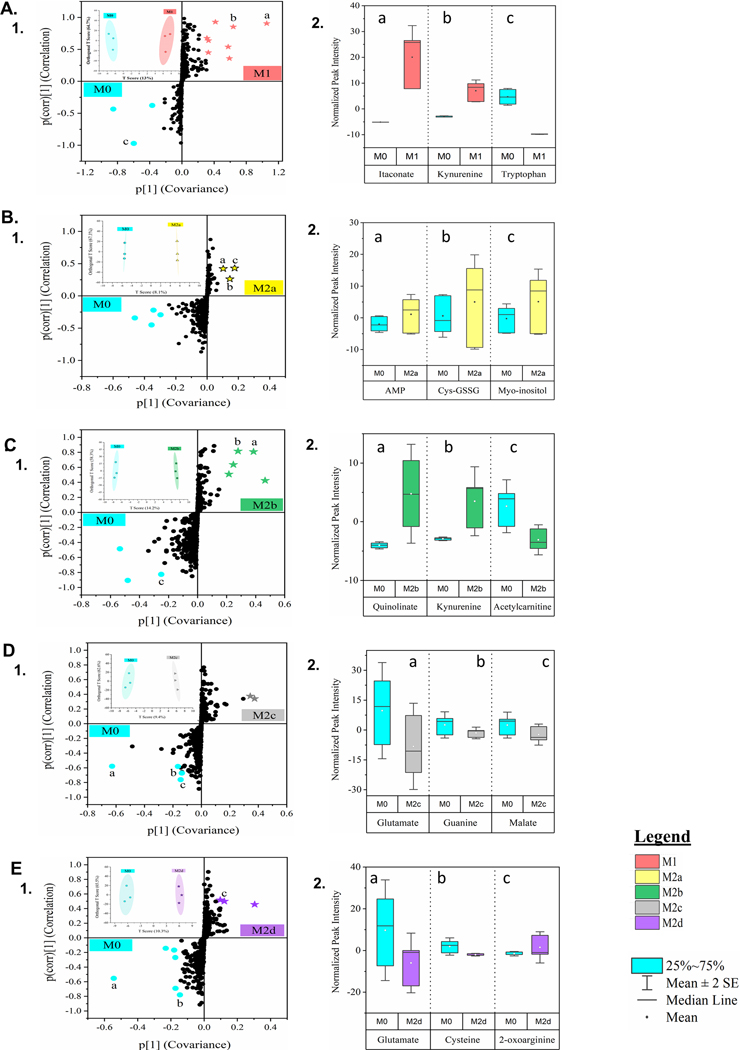
Figure 5: Untargeted Metabolomics of Five Polarized MΦ Phenotypes Identifies Biomarkers of Shared and Unique Profile Relative to the Parent, Resting MΦ Phenotype.
Global metabolomics profile of the parent, resting MΦ phenotype (M0) and five polarized MΦ phenotypes identified 498 compounds of known identity, normalized to cell count. MΦ metabotype clustering was determined through Orthogonal Projections to Latent Structures Discriminant Analysis (OPLSDA), inset clustering scores plots with 2D T-scores (A.1, B.1, C.1, D.1, & E.1). S-plot comparison of each polarized MΦ phenotype was plotted relative to the resting, parent MΦ phenotype (denoted in blue) (A.1, B.1, C.1, D.1, & E.1) and normalized peak intensity for biomarkers of particular note (a, b, c) are displayed as box-whisker plots (A.2, B.2, C.2, D.2, & E.2). Multivariate, cluster analysis is displayed for all five polarized MΦ phenotypes: M1 MΦs (IFN-γ/LPS treated, shown in red) (A), M2a MΦs (IL-4/IL-13 treated, shown in yellow) (B), M2b MΦs (IC/LPS treated, shown in green) (C), M2c MΦs (IL-10 treated, shown in gray) (D), and M2d MΦs (IL-6/LIF treated, shown in purple) (E). Untargeted metabolomics profiles are based on three experimental replicates for each MΦ phenotype (N=3).
To identify putative metabolite biomarkers separating each polarized phenotype from the parent M0 MΦs, OPLSDA model contributions from metabolite measurements (covariance; x-axis) and the reliability of the measurements (correlation; y-axis) were projected in S-plots (Fig. 5A.1-E.1). Additionally, biochemically significant metabolites with high covariance (−0.2 ≤ covariance ≥ 0.2), correlation (−0.6 ≤ correlation ≥ 0.6), and predictive value for metabolic modeling [38] are shown for each MΦ phenotype (Fig. 5A.2-E.2). As the generally accepted paradigm contends that MΦ phenotypes fall into the classically activated M1 and alternatively activated M2 subgroups, SUS graphs of the M1 MΦ metabotype compared to each M2 MΦ subtype were employed to identify shared and unique metabolite biomarkers associated with each pairing (Fig. 6A.1-D.1). Associated correlation scores and statistically significant metabolites (FC > 2.0 and p < 0.05) are depicted in the corresponding tables (Fig. 6A.2-D.2) and volcano plots (Fig. 6A.3-D.3), respectivelly.
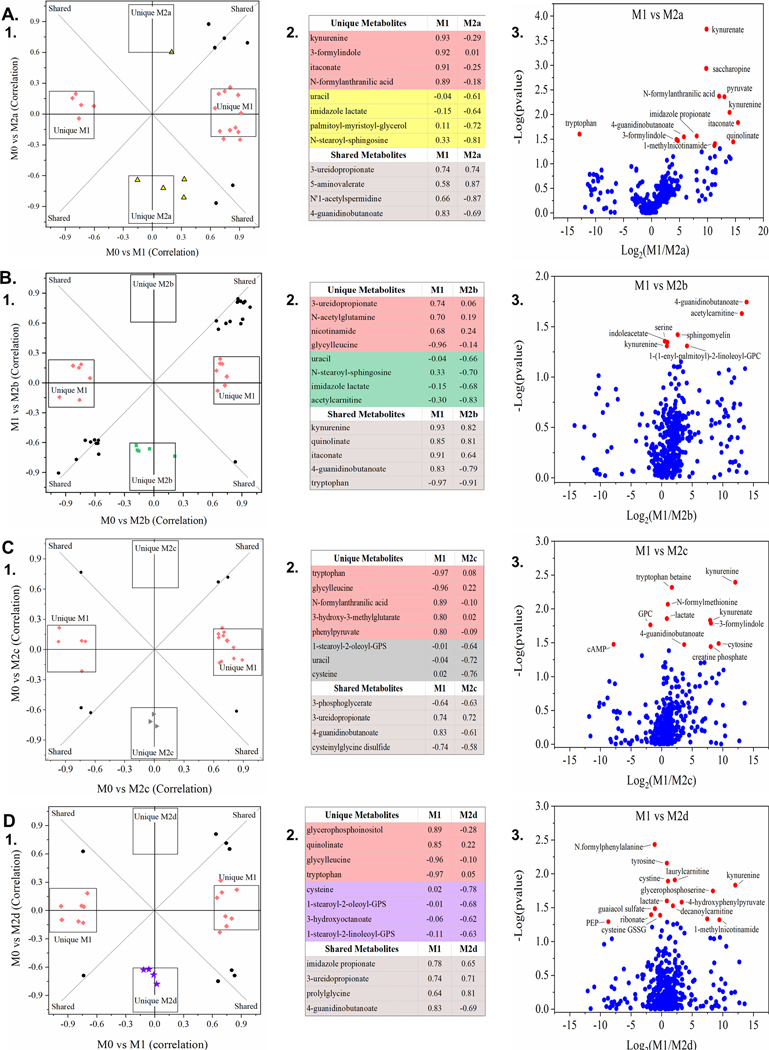
Figure 6: Global Metabolomics Comparison of the Classically Activated MΦ Phenotype (M1 MΦ) to Four Subtypes of Alternately Activated MΦs (M2a, M2b, M2c, & M2d MΦ).
OPLSDA clustering relative to the parent, resting MΦ phenotype (M0 MΦ) identified metabolite biomarkers correlated with all five polarized MΦ phenotypes based on OPLSDA variable influence on projection (VIP) values. VIP values were used to identified both shared and unique metabolite biomarkers of the classically-activated M1 MΦ phenotype and four subtypes of the alternately-activated M2 MΦ phenotypes, displayed as OPLSDA Shared and Unique Structures (SUS) plots (A.1, B.1, C.1, and D.1). For each SUS plot, metabolite biomarkers uniquely correlated to the M1 MΦ phenotype (red diamonds) are plotted along the x-axis and those metabolite biomarkers uniquely correlated to the M2 MΦ phenotype are plotted along the y-axis. Shared metabolite biomarkers (black circles) are plotted along the diagonals, reflecting biomarker positive/negative correlation to each polarized MΦ phenotype relative to the parent, resting MΦ phenotype (M0 MΦ). Associated biomarkers and correlation scores are shown in matching tables (A.2, B.2, C.2, and D.2). OPLSDA SUS plots are displayed for four polarized M2 MΦ phenotypes, relative to the M1 MΦs (IFN-γ/LPS treated, shown in red): M2a MΦs (IL-4/IL-13 treated, shown in yellow) (A), M2b MΦs (IC/LPS treated, shown in green) (B), M2c MΦs (IL-10 treated, shown in gray) (C), and M2d MΦs (IL-6/LIF treated, shown in purple) (D). Untargeted metabolomics profiles are based on three experimental replicates for each MΦ phenotype (N=3).
Contrary to phenotyping by gene expression and correlated functionality, metabotyping each MΦ group did not result in clustering patterns with M2b MΦs more closely related to M1 MΦs and the rest of the M2 subtypes clustered together; however, the unique and shared metabolites within each comparison differed considerably. Metabolites identified through metabotype comparison between M1 and M2b MΦs (Fig. 5A,C and Fig. 6B) indicated metabolic investment in anti-microbial function with itaconate accumulation (correlation score (p(corr)) = 0.91, 0.64, respectively) and antioxidant capacity evidenced by tryptophan consumption (p(corr) = −0.97, −0.91, respectively), kynurenine accumulation (p(corr) = 0.93), and quinolinate accumulation (p(corr) = 0.85, 0.81, respectively), as previously described [44]. Metabolites involved in glutamate metabolism such as N-acetylglutamine (p(corr) = 0.70, 0.19, respectively) correlated highly with M1 MΦs, while BCAA metabolites such as acetylcarnitine (p(corr) = −0.30, −0.83, respectively) were uniquely correlated to M2b MΦs.
The remaining M2 MΦ subtypes (M2a, M2c, and M2d) cluster separation from the M0 MΦs indicated less metabolic divergence upon polarization (Fig. 5B, D, E); however, unique metabolic features could be distinguished between the subtypes. M2a MΦs accumulated cysteine-GSSG and myo-inositol, metabolites involved with glutathione recycling and had similar correlation with uracil from pyrimidine metabolism as M2b and M2c MΦs (p(corr) = −0.61, −0.66, −0.72, respectively) (Fig. 6A-C). Glutamate was consumed by both M2c and M2d MΦs relative to M1 MΦs (p(corr) = −0.58, −0.55, respectively) (Fig. 5D-E and data not shown). Arginine catabolism was indicated by M2d MΦ accumulation of 2-oxoarginine (Fig. 5E.2) and for all M2 MΦ subtypes as demonstrated by negative correlation scores for 4-guanidinobutanoate in M2a, M2b, M2c, and M2d MΦ (p(corr) = −0.87, −0.79, −0.61, and −0.69, respectively) (Fig. 6A.2-D.2), while histidine metabolism was indicated for M2a and M2b MΦs through negative correlation to imidazole lactate (p(corr) = −0.64, −0.68, respectively) (Fig. 6A.2-B.2). Finally, notable differences in lipid metabolism between the M1 and M2 MΦ phenotypes were exhibited by the accumulation of fatty acids and their derivatives in M1 MΦs and corresponding negative correlations within the M2 MΦ subtypes (Fig. 6B.2-D.2; Supplemental Fig. 4B-F).
Metabolic Pathway Analysis to Defined Unique Metabotypes for Each MΦ Polarized Phenotype
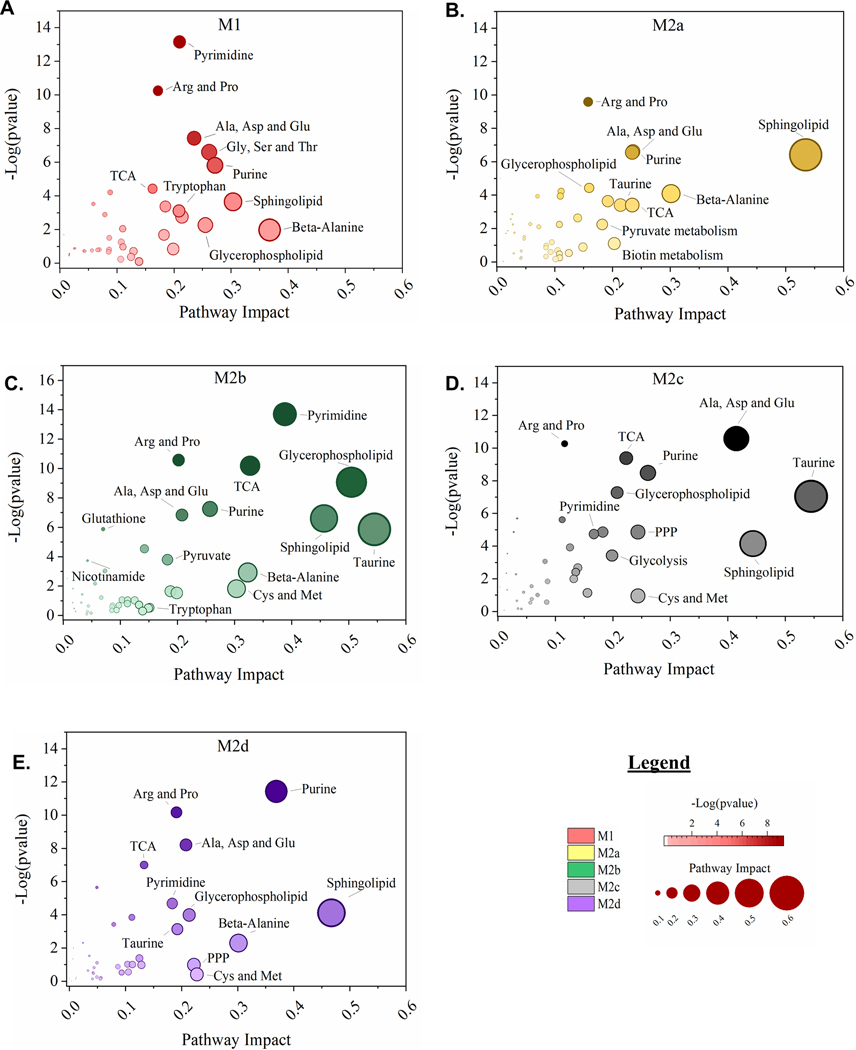
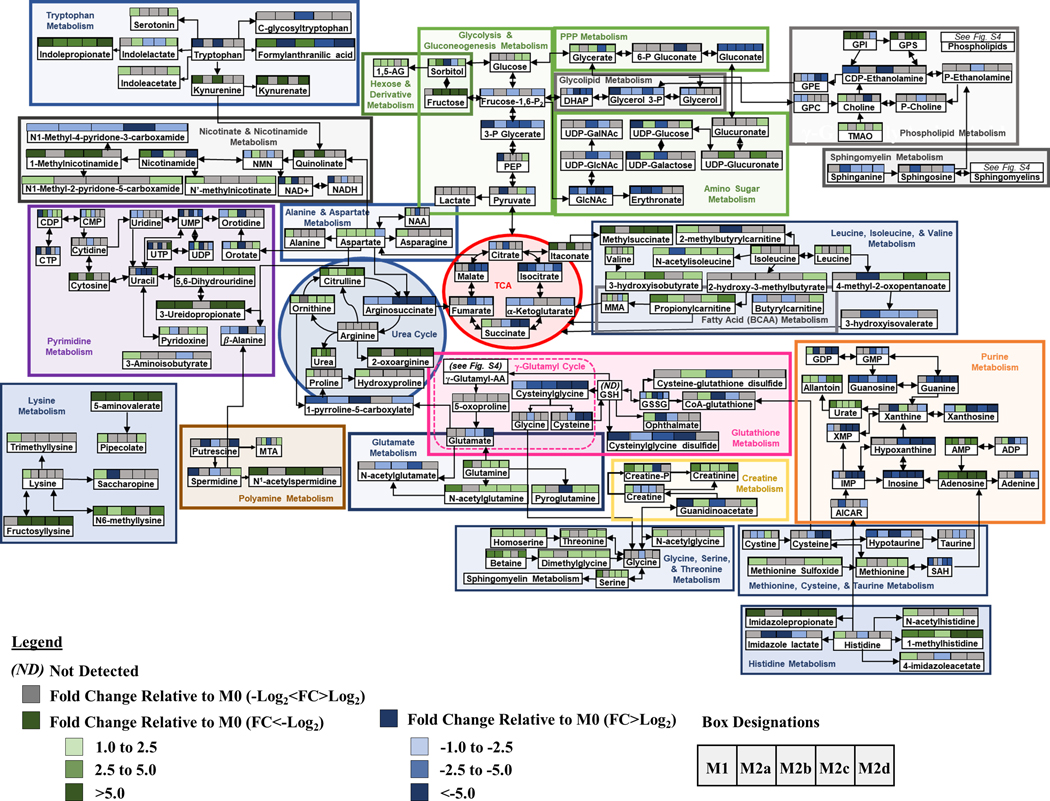
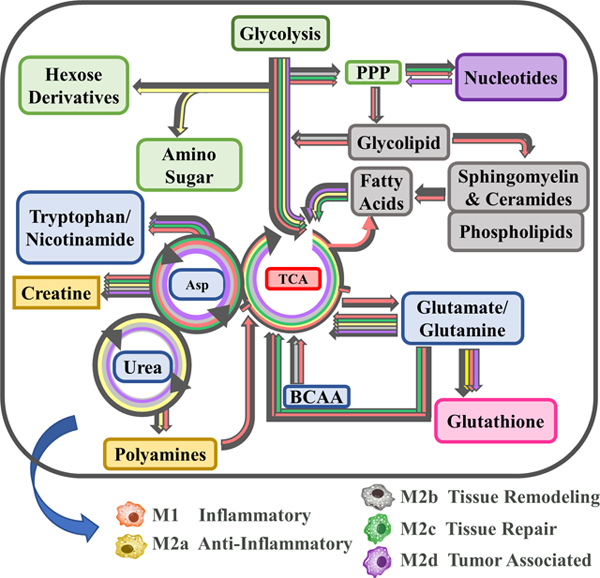
Using topological mapping of global metabolomics organized by p-value from pathway enrichment analysis (y-axis) relative to impact score from pathway topology analysis (x-axis), metabotype was further characterized for each polarized MΦ phenotype and provided a global view of metabolic impact from polarization. For each node, color intensity and size illustrate p-value significance and pathway impact, respectively (Fig. 7). To further support metabotyping by global metabolomics pathway analysis, metabolism-related SDEGs up- or down-regulated relative to the resting parent M0 MΦs are displayed (green or blue, respectively) (Fig. 8). Finally, a comprehensive summary of this metabolic analysis is illustrated in Figure 9. Each colored box above the listed metabolite represents significant fold change relative to the parent M0 MΦs (1 < Log2 < −1). Numerical values corresponding to the color scale are included in Supplemental Table 2.
Figure 7: MΦ Polarization into Functional Phenotypes Activates Distinct Metabolic Pathways.
Untargeted, global metabolomics profile of polarized MΦ phenotypes identified 498 known compounds, normalized to total cell count, and profiled relative to the resting M0 MΦ phenotype. For normalized, M0-relative metabolite profiles, over-representation analysis by hypergeometric testing and pathway topology analysis by two node centrality measures (degree centrality and betweenness centrality) was performed for each polarized MΦ phenotype including M1 MΦs (IFN-γ/LPS treated, shown in red) (A), M2a MΦs (IL-4/IL-13 treated, shown in yellow) (B), M2b MΦs (IC/LPS treated, shown in green) (C), M2c MΦs (IL-10 treated, shown in gray) (D), and M2d MΦs (IL-6/LIF treated, shown in purple) (E). Metabolic Pathway Topology is plotted as Pathway Impact (x-axis) and significance of pathway topography (-Log [p value], y-axis) for each polarized MΦ phenotype.
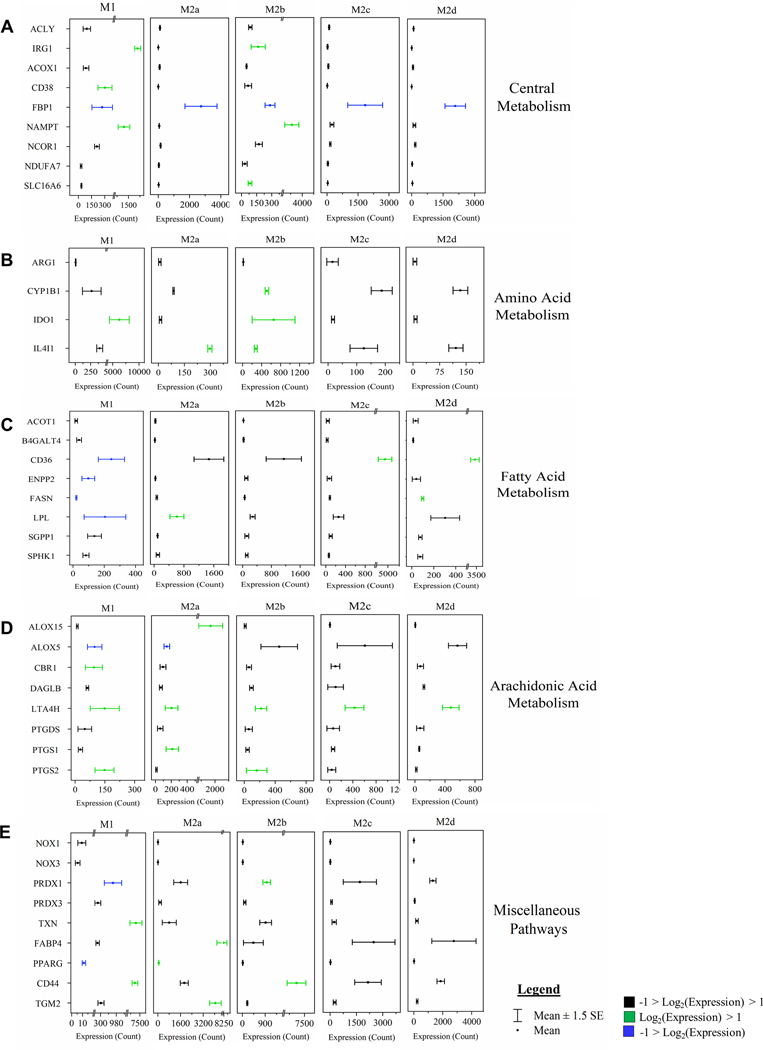
Figure 8: MΦ Polarization into Functional Phenotypes Activates Distinct Metabolic Gene Expression Profiles Relative to the Parent, Resting MΦ Phenotype (M0).
Gene set enrichment analysis (GSEA) and pathway impact scoring of global myeloid gene expression demonstrates distinct patterns in metabolism-related transcripts in the five polarized MΦ phenotypes. Associated metabolism gene expression profiles for most impacted pathways are shown including central metabolism (A), amino acid metabolism (B), fatty acid metabolism (C), arachidonic acid metabolism (D), and miscellaneous pathways (E). Red bars represent gene targets that are differentially regulated (p < 0.05) when compared to the M0 phenotype. Myeloid gene expression was directly detected through molecular barcode probes and normalized to the geometric mean of the housekeeping gene set. Gene expression is mean ± SEM. Pathway topology and metabolic gene expression profiles are based on triplicate, experimental replicates (N=3).
Figure 9: Schematic Demonstrating Metabolic Pathway Flux Relative to the Parent, Resting MΦ Phenotype (M0).
Metabolic flux of selected metabolites within the specified pathways are denoted by arrow direction. Each of the colored boxes above the listed metabolite represents fold change relative to the parent M0 MΦs (1 < Log2 < −1). Fold change values (-Log2 < FC > Log2) are denoted in gray. Fold change values (-Log2 < FC) are denoted in shades of green and fold change values (FC > Log2) are denoted in gray shades of blue.
As MΦs are well characterized to be glycolytic, it was not surprising that FBP1, the gene encoding the FBPI regulator of gluconeogenesis, was significantly down regulated in all the polarized MΦs, especially M1 and M2b MΦs (Fig. 8A). Additionally, the accumulation of glucose observed in the M2c and M2d MΦs (Fig. 9: Glycolysis & Gluconeogenesis) suggests phenotypic variation in the utilization of glucose in MΦ immunometabolism. The accumulation of glycerate (Fig. 9: PPP) in the M1, M2b, and M2d MΦs is suggestive of flux into the PPP for each of these phenotypes. From the topography analysis, the most statistically significant pathway observed was pyrimidine metabolism for both M1 and M2b MΦs (Fig. 7A & C; Fig. 9) while the purine pathway was most prevalent for M2d (Fig. 7E; Fig. 9). Furthermore, the upregulation of thioredoxin (encoded by TXN), a redox mediator, in M1 MΦs and the CD44 transcript in M1 and M2b MΦs (Fig. 8A and 8D), known to enhance glycolysis and metabolic flux into the PPP [45], reinforced this pathway for immunometabolism in M1 and M2b MΦs.
One established metabotype feature of M1 polarization is the decoupled TCA after citrate and succinate, marked by accumulation of itaconate and succinate [46], which was observed in the M1 polarized MΦs (Fig. 9). Moreover, the upregulation of genes encoding an itaconate regulator, IRG1 (Fig. 8A), and succinate-stabilized HIF-1 transcript (Supplemental Table 1) is consistent with these findings. Interestingly, M2b MΦs also exhibited similar IRG1 and HIF-1 upregulation (Fig. 8A) along with itaconate accumulation; however, these MΦs did not accumulate succinate, contrasting with M1 MΦs. The remaining M2 MΦ subtypes (M2a, M2c, and M2d) all demonstrated metabolic flux patterns consistent with an intact TCA (Fig. 7–9).
For amino acid metabolism, the Arg & Pro and Ala, Asp, & Glu metabolic pathways were similarly impacted by polarization (p < 0.05) for all the MΦ phenotypes. In M1 MΦs, both glutamine and aspartate have been shown to replenish TCA metabolites such as α-ketoglutarate and fumarate, presumably depleted as a result of a decoupled TCA [26], and steady-state levels of both fumarate and malate were observed in the M1 MΦ, as well as accumulation of glutamine to feed into the TCA (Fig. 9). While only M1 MΦs accumulated alanine and asparagine, aspartate was accumulated in all polarized phenotypes apart from M2d MΦs (Fig.9). Aspartate accumulation can drive β-alanine metabolism, which was highly impacted for the M1, M2a, M2b, and M2d MΦs (Fig. 7A, B, C, and E) and was consumed within these phenotypes during polarization (Supplemental Table 2 and Fig. 9: Pyrimidine). Aspartate can also lead to quinolinate production, as observed for M1 and M2b MΦs (Fig. 9: Nicotinate & Nicotinamide). Additionally, the upregulation of CD38, IDO1, and NAMPT (Fig. 8A & B) coupled with the consumption of tryptophan and accumulation of kynurenine (Fig. 9) reaffirmed previously findings highlighting the importance of tryptophan and nicotinamide metabolism within the M1 and M2b MΦs. In all M2 MΦ subtypes, taurine metabolism was impacted to various degrees during polarization, while cysteine and methionine metabolism were impacted in M2b, M2c, and M2d MΦ (Fig. 7B – E). Cysteine, taurine, and hypotaurine were generally consumed for the M2 MΦ subtypes (Fig. 9), with cysteine potentially feeding directly into glutathione metabolism in response to oxidative stress (Fig. 9). Moreover, diminished levels of oxidized glutathione and γ-glutamylamino acids, produced as a part of the γ-glutamyl cycle, further indicate oxidative stress associated with polarization across phenotypes (Fig. 9; Supplemental Fig. 4A). Finally, Gly, Ser, and Thr metabolism was only identified as significant within the M1 MΦs (Fig. 7A), supporting previous findings in LPS-stimulated MΦs within mice [47].
Metabotyping in all five polarized MΦ phenotypes identified arginine metabolism as both impacted and significant (Fig. 7A-E). While intracellular stores of arginine did not appear to change with polarization, significant changes to metabolites upstream and downstream of arginine did, including arginosuccinate which feeds into the TCA Cycle, 1-pyrroline-5-carboxylate which derives from the γ-glutamyl cycle, and ornithine and citrulline of the Arg-Cit Cycle (Fig. 9: Urea Cycle). Interestingly, despite extensive literature around the expression of arginase (Arg1) as a marker of alternatively activated M2 MΦs, in our model expression of this gene was not significantly upregulated relative to the parent M0 MΦ phenotype (Fig. 8B); however, accumulation of ornithine in M1, M2a, M2c, and M2d MΦs and urea in M1, M2a, M2b, and M2c MΦs suggest this enzyme is activated upon polarization (Fig. 9). Downstream of 1-pyrroline-5-carboxylate, proline and hydroxyproline accumulated only in M1 MΦs, suggesting that the M2 MΦ subtypes may utilize these metabolites for collagen biosynthesis (Fig. 9) [48]. Finally, arginine can be diverted to synthesize creatine through guanidinoacetate, which is highly consumed in both the M1 and M2b MΦs (Fig. 9) and has been shown to suppress IFN-γ responses while facilitating IL-4 polarization in MΦs [49].
Other pathways of interest included pyrimidine and purine metabolism (Fig. 7A-E). In general, these metabolic pathways were highly consumed across all MΦ phenotypes, but notable in the M1 and M2b MΦ consumption of dioxy- and trioxy-pyrimidines (Fig. 9). An offshoot of pyrimidine metabolism through β-alanine, polyamine metabolism was also markedly impacted with consumption of spermidine and accumulation of N1-acetyl-spermidine in both M1 and M2b MΦs (Fig. 9). Hexosamine metabolism displayed a similar pattern with regards to M1 and M2b MΦ consumption; however, the other M2 MΦ subtypes displayed metabolite accumulation, specifically of UDP-glucose, UDP-galactose, and UDP-glucuronate (Fig. 9). Whether these metabolites drive tissue regeneration/repair or act as signaling molecules within each MΦ phenotype, remains to be determined.
Finally, lipid metabolism was impacted in all five polarized MΦs, especially glycerophospholipid and sphingolipid metabolism (Fig. 7A-E). The COX-2 (PTGS2) transcript, that encodes the enzyme involved in the conversion of arachidonic acid to the pro-inflammatory precursor prostaglandin H2, was upregulated in M1 and M2b MΦs, while the transcript ALOX5, associated with anti-inflammatory responses, were downregulated in M1 MΦs (Fig. 8D). Of the other alternatively activated M2 MΦ phenotypes, M2a MΦs upregulated ALOX15, which facilities activation of known M2a MΦ regulators (PPARG, MRC1, and CD36) (Figure 8C, E, and Supplemental Table 1) [50, 51]. Additionally, the FA metabolism associated transcripts CD36, ENPP2, FASN, and LPL were downregulated in M1 MΦs yet upregulated in M2a, M2c, and M2d MΦs to various levels (Fig. 8C). Lipid metabolite profiles reflected this distinction for the M1 MΦ metabotype, with general patterns of lipid accumulation for M1 MΦs and consumption of lipids for the M2 MΦ metabotypes, with the exception of DHA known to have anti-inflammatory properties (Supplemental Fig. 4B-F) [52]. Interestingly, the most dominant MΦ phenotype with regard to FA metabolism was the M2b MΦ within which levels of lipid consumption generally dwarfed all other phenotypes (Supplemental Fig. 4B-F). What impact this difference in lipid metabolism has on biological systems is under active investigation.
MΦ Effector Functions and Metabotype Dynamics Over Time
Notorious for their plasticity, MΦ are rarely profiled over longer periods of stimulus and the ex vivo MΦ polarization model presented herein provided an excellent platform to compare polarization impact at different timepoints. To evaluate temporal shifts in both MΦ effector function and metabotype, polarized MΦ were harvested at both 24- and 72-hours polarization. Within the inflammatory protein subgroup, TNFα, CCL3, and CXCL8 exhibited minor, but not significant, increases in M1 MΦs at 72 hours (Fig. 10A). While M1 MΦs significantly increased IL-12p70 and significantly decreased IP-10 production between the two time points, IFNα production was significantly decreased for M1, M2b, and M2c MΦ phenotypes (p < 0.05; Fig. 10A). Among the inflammation regulating/tissue repair proteins (p < 0.05; Fig. 10B) and growth factors (Fig. 10C), IL-10 and IL-6 production was significantly decreased from 24 to 72 hours polarization for both the M1 and M2b MΦ phenotypes. Interestingly, IL-1β concentrations increased significantly for both phenotypes, while TGF-β concentrations decreased for all polarized MΦ phenotypes (p < 0.05; Fig. 10B).
Figure 10: MΦ Functional Phenotyping at 72 Hours Post Polarization by Gene and Protein Expression of Immunomodulatory Factors.
Multiplex detection of immunomodulatory factors in six MΦ phenotypes harvested at 72 hours were detected using magnetic bead-based quantification of mRNA and secreted protein. Immunomodulatory function includes pro-inflammatory (TNF⍺, IL-12p70, CCL3, CXCL10, CXCL8, and IFN⍺) (A), immune-regulatory/tissue-repair (IL-10, IL-6, IL-1b, IL-1⍺, TGF-b, and CCL2) (B), and growth factors (GM-CSF, PDGF-BB, and VEGF-A) (C) for the M0 resting MΦs (shown in blue), M1 MΦs (IFN-γ/LPS treated, shown in red), M2a MΦs (IL-4/IL-13 treated, shown in yellow), M2b MΦs (IC/LPS treated, shown in green), M2c MΦs (IL-10 treated, shown in gray), and M2d MΦs (IL-6/LIF treated, shown in purple). Diamond-whisker plots display 25%−75% quartile range, median, and mean. Bar charts indicate mean ± SEM. Expression profiles were normalized to total cell count and include three biological replicates (N=3). Statistical analysis was performed for the MFI values (histogram bars) using repeated measures analysis of variance and model-based means post hoc test (p < 0.05) with an asterick denoting statistical significance.
Additionally, GO analysis demonstrated differentially expressed biological pathways between the 24- and 72-hour time points for all polarized phenotypes. M1 MΦs displayed a strong inflammatory signature at 24 hours, which was sustained at 72 hours (Fig. 11A, light red bars); however, by 72 hours, the M1 MΦs had initiated cellular protective/death processes such as the prostaglandin-endoperoxide synthase, negative regulation of apoptosis, and programmed necrotic cell death GO processes (Fig. 11A, dark red bars). For the M2a MΦs, GO processes at 24 hours heavily favored cellular proliferation, ECM remodeling, and angiogenesis (Fig. 11B, light yellow bars). While these cells still exhibited a potential for angiogenesis and ECM remodeling at 72 hours, the upregulation of the cyclooxygenase, leukotriene, histamine, and IFN-α processes at 72 hours were consistent with activation of anti-inflammatory function, viral inhibition, and the pathogenesis of asthma (Fig. 11B, dark yellow bar) [53]. As discussed above, M2b MΦs share a strong inflammatory functional signature with M1 MΦs at 24 hours (Fig. 11C, light green bars); however, by 72 hours, the M2b MΦs had upregulated genetic programs primarily associated with metabolism (Fig. 11C, dark green bars). ECM remodeling, angiogenesis, and efferocytosis functionality associated with M2c MΦs remained relatively consistent across both time points, with the interesting exception of IL-23 and IL-18 regulation, which are both generally associated with pro-inflammatory functionality. Finally, M2d MΦ functionality at 72 hours persisted with the angiogenic, profibrotic, and anti-inflammatory potential (Fig. 11D, light and dark purple bars); however, lipid metabolism was uniquely profiled at 72 hours, raising the interesting question of how this might translate to immunomodulation in this M2 MΦ subtype.
Figure 11: Gene Expression Profile Comparison of Polarized MΦ Phenotypes Identifies Unique and Overlapping Immunomodulatory Functions between 24- and 72-hour Polarization Times.
Gene Ontology (GO) annotation of global myeloid gene expression identifies the differential biological functionality between the 24 (light colored bars) and 72-hour (dark colored bars) time points in five MΦ phenotypes. For each MΦ phenotype, the top 15 impacted GO processes significantly enriched (p < 0.05) for both time points were plotted as such: M1 MΦs (IFN-γ/LPS treated, shown in red) (A), M2a MΦs (IL-4/IL-13 treated, shown in yellow) (B), M2b MΦs (IC/LPS treated, shown in green) (C), M2c MΦs (IL-10 treated, shown in gray) (D), and M2d MΦs (IL-6/LIF treated, shown in purple) (E). GO process annotation was based on three experimental replicates for each MΦ s phenotype (N=3).
As described above, the top 15 impacted metabolic pathway scores were utilized to generate radar plots comparing 24- and 72-hour metabotypes in all five polarized MΦs (Fig. 12A-E, hatched and solid graphs for 24 and 72 hours, respectively). All phenotypes demonstrated significant shifts in metabolism from 24- to 72-hours polarization. For M1, M2a, M2b, and M2c MΦs, glycolysis and the PPP remained a significant part of the metabotype profile at 72 hours; however, glycolysis was significantly increased in the M2a MΦs (Fig. 11B), while remaining consistent in all of the other phenotypes. PPP metabolism also increased in the M1, M2a and M2b MΦs, but decreased in M2c MΦs. Additionally, amino acid and lipid metabolism was significantly enhanced for both the M1 and M2b MΦs at 72 hours. The global differences in metabolic impact between the M2a (Fig. 11B) and M2c (Fig. 11D) MΦs is quite interesting in that they demonstrated nearly opposite metabotype profiles for the two time points. For example, M2a MΦs exhibited higher impact scores for several amino acids, including BCAA, as well as sphingolipid, galactose, glutathione, and purine metabolism at 24 hours (Fig. 11B, hatched plot), whereas these same pathways were more highly impacted at 72 hours for M2c MΦs (Fig. 11D, solid plot). Another interesting comparison is the different shifts in galactose, glutathione, purine, and pyrimidine metabolism in the M1 and M2b MΦs (Fig. 11A & C), further supporting the distinct metabotype profiles of these phenotypes despite their similar inflammatory signatures. Finally, in contrast to M1, M2a, and M2b MΦ metabolic dynamics, from 24 hours to 72 hours, M2d MΦs undergo a significant constriction in metabolic activity (Fig. 11E). This is especially evident when compared to the M2b MΦs, which significantly expand metabolic activity at 72 hours (Fig. 11C). While these observations in the dynamics of MΦ functionality and metabotype over time demonstrate that each of these MΦ phenotypes can exhibit significant plasticity, the causal relationship between metabolism and biological function remains an expansive area for future investigations.
Figure 12: Metabotype Profiling of Polarized MΦs at 24 and 72 Hours.
Pathway impact scores derived from pathway topology analysis were compared at 24 and 72 hours for the M1 MΦs (IFN-γ/LPS treated, shown in red) (A), M2a MΦs (IL-4/IL-13 treated, shown in yellow) (B), M2b MΦs (IC/LPS treated, shown in green) (C), M2c MΦs (IL-10 treated, shown in gray) (D), and M2d MΦs (IL-6/LIF treated, shown in purple) (E). The top 15 enriched pathways are depicted in each spider plot with the hatched plots and solid plots representing the 24- and 72-hour time points, respectively.
DISCUSSION
Previous studies have described multiple MΦ phenotypes [13, 15, 19, 43]; however, to our knowledge, this study represents the most comprehensive functional phenotyping and correlated metabotype of the current MΦ phenotypes described. MΦ plasticity is uniquely dynamic and the correlated profiling of metabotype and functional phenotype presented herein provides a framework for further investigation into how this dynamic plasticity is driven through integration between immune function and metabolism. While there are multiple lines of further investigation indicated by this study, there are specific correlations between metabolism and functional phenotype that support the overall hypothesis of metabolic immunomodulation driving MΦ plasticity in each phenotype.
Exposure of resting M0 MΦs to IFNγ and LPS resulted in the classically defined M1 MΦ phenotype characterized by a strong, proinflammatory signature as evidenced by CD40highCD86highCD163low display, the secretion of several inflammatory cytokines and chemokines, the upregulation of M1-associated transcripts, and the functional profile identified through GO process analysis. Distinct metabolic shifts were also identified and supported previous findings. Analysis of our metabolic data supports the hypothesis that reprogramming in this MΦ phenotype from OXPHOS to aerobic glycolysis supports rapid ATP production and the provision of several glycolytic intermediates such as PKM2, GAPDH, and enolase for IL-1α/β, IL-6, and TNFα production [25, 54–56]. In addition, metabotyping supports previous reports of a decoupled TCA cycle in M1 MΦs [46]. The first break in the TCA cycle results in an accumulation of citrate, which is diverted for itaconate production through upregulation of IRG1. In addition to antimicrobial activity [57], itaconate serves as an inhibitor of SDH, resulting in the accumulation of succinate and a second observed break in the TCA cycle, as shown in the M1 MΦ metabotype and as previously reported [46, 57]. The accumulated succinate enhances HIF-1α stabilization and the production of IL-1β [23, 57, 58]. TCA cycle disruption also results in de novo FAS through excess citrate [10] and lipid accumulation was observed in the M1 MΦ metabotype, shown to be essential for the synthesis of inflammatory mediators [59]. For example, the arachidonic metabolism of the M1 MΦ metabotype is shown to drive prostaglandin synthesis [60]. Overall, the decoupled TCA cycle and downstream metabolic outcomes of M1 MΦ polarization shown herein support the previously demonstrated metabolic immunomodulation of this well-described functional phenotype.
Within the context of wound healing, the most well described M2 MΦ subset is the alternately-activated, anti-inflammatory M2a MΦ, polarized via exposure to IL-4/IL-13 [61]. M2a MΦs upregulate and secrete numerous biological factors, such as IL-1RA to suppress IL-1 mediated inflammation [16], the chemokine ligands CCL3, CCL17, CCL18, and CCL22, and growth factors GM-CSF, TGF-β, and PDGF needed to promote the recruitment, and differentiation of numerous cell types necessary for the proliferative phase of wound healing [62]. In our ex vivo model, M2a MΦs upregulated cell-surface MRC1, which facilitates the clearance of glycosylated pathogens, inflammatory proteins, and the efferocytosis of apoptotic cells in a CD36-dependent fashion [63, 64]. Additionally, MRC1 is known to target mannosylated antigen intracellularly for MHC-II compartments and antigen presentation in dendritic cells [65]. In support of an APC role, the polarized M2a MΦs also increased expression of the MHC-II receptor HLA-DR. Metabolically, M2a MΦs exibit both active glycolysis and an intact TCA cycle. In constrast to previous research [28], decreased pools of lipids indicated active FAO; however, CD36-dependent FAO is critical for M2a functionality at later time points [28]. Supporting Jha, et al., (2015), who demonstrated the necessity of glutamine for amino and nucleotide sugar metabolism and M2a polarization markers CD301 and MRC1 [26], we also observe increased metabolic activity within the hexosamine biosynthesis pathway and increased pools of UDP-GlcNAc-associated modules.
While the majority of research on MΦ polarization has focused on classically-activated, pro-inflammatory M1 MΦs and alternately-activated, anti-inflammatory M2a MΦs, other M2-associated phenotypes have been reported in the literature with functionality distinct from that of M2a MΦs. For example, resting MΦs exposed to TLR-agonist, endogenous ligands coupled with FCγ receptor ligation resulted in a polarized phenotype secreting pro-inflammatory cytokines while promoting biological effector mechanisms associated with the M2 family of MΦs [19, 66], referred to as M2b MΦs. Observed to be ineffective at bacterial clearance [67], M2b MΦs display characteristics indicative of a tissue remodeling functional phenotype [68]. Induced herein via combined LPS and IC polarization, the M2b MΦs profiled as distinct from any other M2 MΦ subtype by secreting proinflammatory cytokines TNFα, IL-1β, and IL-6 and upregulating transcripts that promote Th17 responses, including TNF, IL-1β, IL-23A, IL-10, CCL1, and CCL5 [9]. Metabolically, M2b MΦs are similar to M1 MΦs with active glycolysis, tryptophan, amino sugar, and pyrimidine metabolism; however, in contrast to M1 MΦs, the TCA cycle appears to be intact in M2b MΦs, as shown by the lack of succinate accumulation. In LPS-activated MΦs, HIF-1α stabilization and IL-1β production has been linked to succinate accumulation via glutamine-dependent anaplerosis or the γ-aminobutyric acid (GABA) shunt [23]; however, this does not appear to be an active mechanism in M2b MΦs and thus warrants further investigation. Additionally, the M2b MΦ lipid metabolite profile suggests active FAO or OXPHOS similar to the other M2 subtypes.
The M2c MΦ functional phenotype is characterized by CD163 and is induced through IL-10 stimuli, as utilized herein [20]; therefore, it was not surprising that cell-surface marker analysis characterized this M2 MΦ subtype as CD86lowCD163high. These findings, along with the observed upregulation of MerTk, a key component of efferocytosis, is consistent with phagocytosis GO processes being profiled. CD163, the hemoglobin scavenger receptor, mediates Hb-oxidative tissue damage following hemolysis and induces the expression of the anti-inflammatory enzyme heme oxidase 1 (HMOX1; data not shown) [69]. Beyond CD163, functional phenotyping of the M2c MΦs is consistent with previous phenotypic characterization of inflammatory regulation, mediation of vascular insult, and reduction of oxidative stress [14, 69, 70]. To date, extraordinarily little is known regarding the metabolism of M2c MΦs and has been limited to carbohydrate and glutamate metabolism [24]. In contrast to previous reports by Rodriguez-Prados et al. [24], M2c MΦs accumulated a significant amount of glucose and diminished pools of glutamate in comparison to the M0 parent cells. Given the necessity of glutamate in ECM remodeling, it is probable that glutamate is being diverted to collagen synthesis (as reviewed in Karna, et al. 2020 [71]). In addition to active glycolysis, an observed shift to the PPP via glucuronate and the accumulation of UDP-glucuronate suggests a potential investment in the synthesis of hyaluronic acid for tissue regeneration and anti-inflammatory responses during wound healing [72].
Given the remarkable plasticity of MΦs and the equally disparate array of tumor-specific microenvironments, it is not surprising that TAMs, as a collective, exhibit diverse functionality resembling both M1 and M2 polarized phenotypes [73]. As such, TAMs have been associated with protein expression patterns observed in both acute wounding (i.e. IL-6, and IL-1β) and wound resolution (i.e. TGF-β, PDGF and VEGFA) [22]. IL-6 appears to be central to many of the underlying processes that drive TAM functionality and was utilized to polarize the M2d MΦs. In support of previous reports, the M2d MΦ transcriptional profile included upregulation of HMOX1, promotion of angiogenesis, and immunosupression [22] [74] [21]. Furthermore, TGF-β and PDGF-BB, secreted at significant levels in these cells, are well-documented factors associated with tumor progression and generally predict poor outcomes [75]. While the tumor metabolic environment typically has some degree of hypoxia, in our ex vivo model, M2d MΦs were not cultured under hypoxic conditions and glucose accumulation was observed, indicating functional glycolysis, as has been observed in primary tumors [4, 76, 77]. The tumor environment has also been characterized as abundant in lipids and M2d MΦs were observed to highly express CD36, the fatty acid translocase integral membrane protein, as well as significant FAO.
Finally, Gabriel, et al. (2014) demonstrated differences in the timing of gene expression during MΦ polarization and proposed that this temporal polarization may distinguish MΦ phenotypes in vivo [78]. To investigate temporal effects on polarization, we extended our ex vivo MΦ polarization model out to 72 hours. While pro-inflammatory MΦs (M1) shifted to programmed cell death functions and anti-inflammatory MΦs (M2a and M2c) continued to promote tissue remodeling/repair function, M2b and M2d MΦs demonstrated significant and opposite shifts in metabolic programming. For example, apart from glycerophospholipid metabolism and BCAA degradation, the M2d MΦs displayed a massive contraction of metabolic activity, while the M2b MΦs display a massive increase in metabolic activity. The functional consequences of this metabolic shift were accompanied by down regulation of TGFβ in M2d MΦs and up regulation of IL-1β in M2b MΦs; however, further investigation is warranted and ongoing.
Understanding how global metabolic shifts contribute to MΦ plasticity provides for greater insight into mechanisms of wound healing and a foundation for novel innovations in therapeutic design. While much of what has been previously reported has been done using murine models or immortablized cell lines, the metabolic profiles of murine-derived MΦs compared to primary human-derived MΦs are inconsistent, demonstrating how much is still unknown about how metabolism drives function in these diverse MΦ phenotypes [79]. This study presents our ex vivo MΦ model system utilizing primary, human blood-derived MΦs, allowing nearly homogeneous polarization of MΦ phenotypes without restriction of plasticity, as seen in in vitro models with immortalized cell lines. In situ validation of the functional phenotypes and associated metabotypes presented herein is ongoing and outside of the scope of the current study; however, demonstrating the potential in vivo context across numerous tissues and pathologies provides unlimited opportunity for investigation. In addition, these findings demonstrate correlation between MΦ functional phenotype and metabotype, but deciphering the intricate interplay between metabolism and immunomodulation remains an area of active investigation.
Supplementary Material
Acknowledgments
This work was supported in part by the U.S. Department of Veterans Affairs, Office of Research and Development Biomedical Laboratory Research Program, the Idaho INBRE Program (NIH NIGMS P20 GM103408), a NIAID-NIH award (RO3AI135998; PI Ammons), and the IVREF Center of Biomedical Research Excellence in Emerging/Reemerging Infectious Diseases (NIH NIGMS P20GM109007). This content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, U.S. Department of Veterans Affairs, or the United States Government.
Abbreviations
- αKG
alpha ketoglutarate
- AHS
autologous human serum
- ALA
alanine
- AMP
adenosine monophosphate
- ANOVA
analysis of variance
- APC
allophycocyanin
- APCs
antigen-presenting cells
- ARG
arginine
- ASP
aspartate
- ATP
adenosine triphosphate
- BCAA
branch chain amino acid
- bFGF
basic fibroblast growth factor
- BSA
bovine serum albumin
- CCL
chemokine (C-C motif) ligand
- CD
cluster of differentiation
- CIT
citruline
- CSV
comma-separated value
- CXCL
chemokine (C-X-C motif) ligand
- CYS
cysteine
- DAMPs
damage-associated molecular patterns
- DGS
directed global significance
- ECM
extracellular matrix
- EGF
epidermal growth factor
- FAO
fatty acid oxidation
- FAs
fatty acids
- FASN
fatty acid synthase
- FDR
false discovery rate
- FGF
fibroblast growth factor
- FITC
fluorescein isothiocyanate
- FMO
fluorescence minus one
- Fcγ
fragment crystallizable gamma region
- GLU
glutamate
- GLY
glycine
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- GO
gene ontology
- GSEA
gene set enrichment analysis
- Hb
hemoglobin
- HC
hierarchical clustering
- HESI-II
heated electrospray ionization
- HIF
hypoxia-inducible factor
- HIS
histidine
- HLA-DR
human leukocyte antigen – DR isotype
- ICs
immune complexes
- IFN-γ
interferon gamma
- IL
interleukin
- IL-1Ra
IL-1 receptor agonist
- IRB
institutional review board
- LIF
leukemia inhibitory factor
- LPS
lipopolysaccharides
- M-CSF
macrophage colony stimulating factor
- MEM
minimum essential media
- MerTK
Mer receptor tyrosine kinase
- MFI
median fluorescence intensity
- MHC-II
major histocompatibility complex class II
- MMP
matrix metalloproteases
- MRC1
mannose receptor
- MΦs
macrophages
- NADPH
nicotinamide adenine dinucleotide phosphate
- NO
nitric oxide
- OPLSDA
orthogonal PLSDA
- OXPHOS
oxidative phosphorylation
- PBMCs
primary blood-derived mononuclear cells
- PBS
phosphate buffered saline
- PCA
principal component analysis
- PDGF
platelet-derived growth factor
- PE
phycoerythrin
- PerCP-Cy
peridinin chlorophyll protein complex cyanine
- PFPA
perfluoropentanoic acid
- PGF
placental growth factor
- PLSDA
partial least-squares to latent structures discriminant analysis
- PPP
pentose phosphate pathway
- PRO
proline
- QC
quality control
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RPMI
Roswell Park Memorial Institute
- RSD
relative standard deviation
- SDEGs
significantly differentially expressed genes
- SDH
succinate dehydrogenase
- SER
serine
- SLE
systemic lupus erythematosus
- SYK
spleen tyrosine kinase
- SPMs
specialized pro-resolving mediators
- SUS
Shared and Unique Structure
- T helper cells
Th cells
- TAMs
tumor associated MΦs
- TCA
tricarboxylic acid cycle
- TGF-β
transforming growth factor-β
- THR
threonine
- TLR
toll-like receptor
- TNF
tumor necrosis factor
- TXN
thioredoxin
- UDS
undirected global significance
- UHPLC/MS/MS
ultra-high-performance liquid chromatography/tandem accurate mass spectrometry
- VEGF
vascular endothelial growth factor
Footnotes
Disclosures
The authors declare no conflicts of interest.
Authorship
C.B.A, M.C.B.A, and M.M.D. conceptualized and designed the experimental approach and supervised sample collection for this research. C.B.A, T.M.W.L., H.L.S., and J.G. contributed equally to the execution of the experiments and data collection. C.B.A and M.C.B.A. performed the data analysis, generated the figures, and wrote the manuscript. All authors participated in the editing process.
References
- 1.Kodelja V, Müller C, Tenorio S, Schebesch C, Orfanos CE, Goerdt S. Differences in angiogenic potential of classically vs alternatively activated macrophages. Immunobiology 1997; 197: 478–493. [DOI] [PubMed] [Google Scholar]
- 2.Stout RD, Jiang C, Matta B, Tietzel I, Watkins SK, Suttles J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. The Journal of Immunology 2005; 175: 342–349. [DOI] [PubMed] [Google Scholar]
- 3.Daley JM, Brancato SK, Thomay AA, Reichner JS, Albina JE The phenotype of murine wound macrophages. Journal of Leukocyte Biology 2010; 87: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penny HL, Sieow JL, Adriani G, Yeap WH, See Chi Ee P, San Luis B, Lee B, Lee T, Mak SY, Ho YS Warburg metabolism in tumor-conditioned macrophages promotes metastasis in human pancreatic ductal adenocarcinoma. Oncoimmunology 2016; 5: e1191731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halbrook CJ, Pontious C, Kovalenko I, Lapienyte L, Dreyer S, Lee H-J, Thurston G, Zhang Y, Lazarus J, Sajjakulnukit P. Macrophage-released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell metabolism 2019; 29: 1390–1399. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao W, Peters JH, Nieman D, Sharma M, Watson T, Yu J. Macrophage subtype predicts lymph node metastasis in oesophageal adenocarcinoma and promotes cancer cell invasion in vitro. British journal of cancer 2015; 113: 738–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbings SL, Goyal R, Desch AN, Leach SM, Prabagar M, Atif SM, Bratton DL, Janssen W, Jakubzick CV Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages. Blood 2015; 126: 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelpoel LTC, Hansen IS, Rispens T, Muller FJM, van Capel TMM, Turina MC, Vos JB, Baeten DLP, Kapsenberg ML, de Jong EC, den Dunnen J. Fc gamma receptor-TLR cross-talk elicits pro-inflammatory cytokine production by human M2 macrophages. Nature communications 2014; 5: 5444–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsanaktsi A, Solomou EE, Liossis S-NC Th1/17 cells, a subset of Th17 cells, are expanded in patients with active systemic lupus erythematosus. Clinical Immunology 2018; 195: 101–106. [DOI] [PubMed] [Google Scholar]
- 10.O’Neill LAJ, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol 2016; 16: 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lachmandas E, Boutens L, Ratter JM, Hijmans A, Hooiveld GJ, Joosten LA, Rodenburg RJ, Fransen JA, Houtkooper RH, Van Crevel R. Microbial stimulation of different Toll-like receptor signalling pathways induces diverse metabolic programmes in human monocytes. Nature microbiology 2017; 2: 16246. [DOI] [PubMed] [Google Scholar]
- 12.Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M, Udalova IA IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nature immunology 2011; 12: 231. [DOI] [PubMed] [Google Scholar]
- 13.Edwards JP, Zhang X, Frauwirth KA, Mosser DM Biochemical and functional characterization of three activated macrophage populations. Journal of Leukocyte Biology 2006; 80: 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lurier EB, Dalton D, Dampier W, Raman P, Nassiri S, Ferraro NM, Rajagopalan R, Sarmady M, Spiller KL Transcriptome analysis of IL-10-stimulated (M2c) macrophages by next-generation sequencing. Immunobiology 2017; 222: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duluc D, Delneste Y, Tan F, Moles M-P, Grimaud L, Lenoir J, Preisser L, Anegon I, Catala L, Ifrah N, Descamps P, Gamelin E, Gascan H, Hebbar M, Jeannin P. Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood 2007; 110: 4319–4330. [DOI] [PubMed] [Google Scholar]
- 16.Fenton MJ, Buras J, Donnelly R. IL-4 reciprocally regulates IL-1 and IL-1 receptor antagonist expression in human monocytes. The Journal of Immunology 1992; 149: 1283–1288. [PubMed] [Google Scholar]
- 17.Jetten N, Verbruggen S, Gijbels MJ, Post MJ, De Winther MPJ, Donners MMPC Anti-inflammatory M2, but not pro-inflammatory M1 macrophages promote angiogenesis in vivo. Angiogenesis 2014; 17: 109–118. [DOI] [PubMed] [Google Scholar]
- 18.Gerber JS and Mosser DM Reversing lipopolysaccharide toxicity by ligating the macrophage Fcγ receptors. The Journal of Immunology 2001; 166: 6861–6868. [DOI] [PubMed] [Google Scholar]
- 19.Ambarus CA, Santegoets KCM, van Bon L, Wenink MH, Tak PP, Radstake TRDJ, Baeten DLP Soluble immune complexes shift the TLR-induced cytokine production of distinct polarized human macrophage subsets towards IL-10. PloS one 2012; 7: e35994–e35994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohlsson SM, Linge CP, Gullstrand B, Lood C, Johansson Å, Ohlsson S, Lundqvist A, Bengtsson AA, Carlsson F, Hellmark T. Serum from patients with systemic vasculitis induces alternatively activated macrophage M2c polarization. Clinical Immunology 2014; 152: 10–19. [DOI] [PubMed] [Google Scholar]
- 21.Hossain F, Al-Khami AA, Wyczechowska D, Hernandez C, Zheng L, Reiss K, Del Valle L, Trillo-Tinoco J, Maj T, Zou W. Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances cancer therapies. Cancer immunology research 2015; 3: 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muliaditan T, Caron J, Okesola M, Opzoomer JW, Kosti P, Georgouli M, Gordon P, Lall S, Kuzeva DM, Pedro L. Macrophages are exploited from an innate wound healing response to facilitate cancer metastasis. Nature communications 2018; 9: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, O’Neill LAJ Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013; 496: 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez-Prados J-C, Través PG, Cuenca J, Rico D, Aragonés J, Martín-Sanz P, Cascante M, Boscá L. Substrate Fate in Activated Macrophages: A Comparison between Innate, Classic, and Alternative Activation. The Journal of Immunology 2010; 185: 605. [DOI] [PubMed] [Google Scholar]
- 25.Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MAR, Sheedy FJ, Gleeson LE, van den Bosch MWM, Quinn SR, Domingo-Fernandez R, Johnston DGW, Jiang J-K, Israelsen WJ, Keane J, Thomas C, Clish C, Vander Heiden M, Xavier RJ, O’Neill LAJ Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell metabolism 2015; 21: 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jha Abhishek K., Huang Stanley C.-C., Sergushichev, Lampropoulou, Ivanova Y, Loginicheva E, Chmielewski K, Stewart Kelly M., Ashall J, Everts B, Pearce Edward J., Driggers Edward M., Artyomov Maxim N. Network Integration of Parallel Metabolic and Transcriptional Data Reveals Metabolic Modules that Regulate Macrophage Polarization. Immunity 2015; 42: 419–430. [DOI] [PubMed] [Google Scholar]
- 27.Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proceedings of the National Academy of Sciences 2013; 110: 7820–7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang SC-C, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O’Neill CM, Yan C, Du H, Abumrad NA, Urban JF Jr., Artyomov MN, Pearce EL, Pearce EJ Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nature immunology 2014; 15: 846–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan Z, Xie N, Cui H, Moellering DR, Abraham E, Thannickal VJ, Liu G. Pyruvate dehydrogenase kinase 1 participates in macrophage polarization via regulating glucose metabolism. The Journal of immunology 2015; 194: 6082–6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malandrino MI, Fucho R, Weber M, Calderon-Dominguez M, Mir JF, Valcarcel L, Escoté X, Gómez-Serrano M, Peral B, Salvadó L. Enhanced fatty acid oxidation in adipocytes and macrophages reduces lipid-induced triglyceride accumulation and inflammation. American Journal of Physiology-Endocrinology and Metabolism 2015; 308: E756–E769. [DOI] [PubMed] [Google Scholar]
- 31.Rath M, Müller I, Kropf P, Closs EI, Munder M. Metabolism via arginase or nitric oxide synthase: two competing arginine pathways in macrophages. Frontiers in immunology 2014; 5: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandra P, He L, Zimmerman M, Yang G, Köster S, Ouimet M, Wang H, Moore KJ, Dartois V, Schilling JD, Philips JA Inhibition of Fatty Acid Oxidation Promotes Macrophage Control of Mycobacterium tuberculosis. mBio 2020; 11: e01139–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders CB, Lawton TMW, Ammons MCB Metabolic immunomodulation of macrophage functional plasticity in nonhealing wounds. Current opinion in infectious diseases 2019; 32: 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomfohr J, Lu J, Kepler TB Pathway level analysis of gene expression using singular value decomposition. BMC bioinformatics 2005; 6: 225–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mudunuri U, Che A, Yi M. and Stephens R.M bioDBnet: the biological database network. Bioinformatics 2009; 25: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang DW, Sherman BT, Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 37.Chong J, Wishart DS, Xia J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Current Protocols in Bioinformatics 2019; 68: e86. [DOI] [PubMed] [Google Scholar]
- 38.Wiklund S, Johansson E, Sjöström L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J. Visualization of GC/TOF-MS-Based Metabolomics Data for Identification of Biochemically Interesting Compounds Using OPLS Class Models. Analytical Chemistry 2008; 80: 115–122. [DOI] [PubMed] [Google Scholar]
- 39.Ambarus CA, Krausz S, van Eijk M, Hamann J, Radstake TRDJ, Reedquist KA, Tak PP, Baeten DLP Systematic validation of specific phenotypic markers for in vitro polarized human macrophages. Journal of Immunological Methods 2012; 375: 196–206. [DOI] [PubMed] [Google Scholar]
- 40.van der Kroef M, Carvalheiro T, Rossato M, de Wit F, Cossu M, Chouri E, Wichers CGK, Bekker CPJ, Beretta L, Vazirpanah N, Trombetta E, Radstake TRDJ, Angiolilli C. CXCL4 triggers monocytes and macrophages to produce PDGF-BB, culminating in fibroblast activation: Implications for systemic sclerosis. Journal of Autoimmunity 2020; 111: 102444. [DOI] [PubMed] [Google Scholar]
- 41.Huang B, Yang X-D, Lamb A, Chen L-F Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell Signal 2010; 22: 1282–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan J-B, Miyauchi-Ishida S, Arimoto K. i., Liu D, Yan M, Liu C-W, Győrffy B, Zhang D-E Type I IFN induces protein ISGylation to enhance cytokine expression and augments colonic inflammation. Proceedings of the National Academy of Sciences 2015; 112: 14313–14318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sudan B, Wacker MA, Wilson ME, Graff JW A systematic approach to identify markers of distinctly activated human macrophages. Frontiers in Immunology 2015; 6: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL Inhibition of T cell proliferation by macrophage tryptophan catabolism. The Journal of experimental medicine 1999; 189: 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamada M, Nagano O, Tateyama S, Ohmura M, Yae T, Ishimoto T, Sugihara E, Onishi N, Yamamoto T, Yanagawa H, Suematsu M, Saya H. Modulation of Glucose Metabolism by CD44 Contributes to Antioxidant Status and Drug Resistance in Cancer Cells. Cancer research 2012; 72: 1438. [DOI] [PubMed] [Google Scholar]
- 46.Cordes T, Wallace M, Michelucci A, Divakaruni AS, Sapcariu SC, Sousa C, Koseki H, Cabrales P, Murphy AN, Hiller K. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. Journal of Biological Chemistry 2016; 291: 14274–14284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishiyama A, Yokote Y, Sakagami H. Changes in Amino Acid Metabolism During Activation of Mouse Macrophage-like Cell Lines. In Vivo 2010; 24: 857–860. [PubMed] [Google Scholar]
- 48.Curran JN, Winter DC, Bouchier‐Hayes D. Biological fate and clinical implications of arginine metabolism in tissue healing. Wound Repair and Regeneration 2006; 14: 376–386. [DOI] [PubMed] [Google Scholar]
- 49.Ji L, Zhao X, Zhang B, Kang L, Song W, Zhao B, Xie W, Chen L, Hu X. Slc6a8-Mediated Creatine Uptake and Accumulation Reprogram Macrophage Polarization via Regulating Cytokine Responses. Immunity 2019; 51: 272–284.e7. [DOI] [PubMed] [Google Scholar]
- 50.Kwon H, Kim S, Kim Y, Lee Y. The contribution of arachidonate 15-lipoxygenase in tissue macrophages to adipose tissue remodeling. Cell death & disease 2016; 7: e2285–e2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lefèvre L, Galès A, Olagnier D, Bernad J, Perez L, Burcelin R, Valentin A, Auwerx J, Pipy B, Coste A. PPARγ ligands switched high fat diet-induced macrophage M2b polarization toward M2a thereby improving intestinal Candida elimination. PloS one 2010; 5: e12828–e12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talamonti E, Pauter AM, Asadi A, Fischer AW, Chiurchiù V, Jacobsson A. Impairment of systemic DHA synthesis affects macrophage plasticity and polarization: implications for DHA supplementation during inflammation. Cellular and Molecular Life Sciences 2017; 74: 2815–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Girodet P-O, Nguyen D, Mancini JD, Hundal M, Zhou X, Israel E, Cernadas M. Alternative macrophage activation is increased in asthma. American journal of respiratory cell and molecular biology 2016; 55: 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie M, Yu Y, Kang R, Zhu S, Yang L, Zeng L, Sun X, Yang M, Billiar TR, Wang H, Cao L, Jiang J, Tang D. PKM2-dependent glycolysis promotes NLRP3 and AIM2 inflammasome activation. Nature Communications 2016; 7: 13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Millet P, Vachharajani V, McPhail L, Yoza B, McCall CE GAPDH binding to TNF-α mRNA contributes to posttranscriptional repression in monocytes: a novel mechanism of communication between inflammation and metabolism. The Journal of Immunology 2016; 196: 2541–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bae S, Kim H, Lee N, Won C, Kim H-R, Hwang Y. i., Song YW, Kang JS, Lee WJ α-Enolase expressed on the surfaces of monocytes and macrophages induces robust synovial inflammation in rheumatoid arthritis. The Journal of Immunology 2012; 189: 365–372. [DOI] [PubMed] [Google Scholar]
- 57.Lampropoulou V, Sergushichev A, Bambouskova M, Nair S, Vincent EE, Loginicheva E, Cervantes-Barragan L, Ma X, Huang SC-C, Griss T. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell metabolism 2016; 24: 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Littlewood-Evans A, Sarret S, Apfel V, Loesle P, Dawson J, Zhang J, Muller A, Tigani B, Kneuer R, Patel S. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. Journal of Experimental Medicine 2016; 213: 1655–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bozza PT, Magalhães KG, Weller PF Leukocyte lipid bodies - Biogenesis and functions in inflammation. Biochimica et biophysica acta 2009; 1791: 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Infantino V, Iacobazzi V, Menga A, Avantaggiati ML, Palmieri F. A key role of the mitochondrial citrate carrier (SLC25A1) in TNFα-and IFNγ-triggered inflammation. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 2014; 1839: 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen L, Wang J, Li S, Yu Z, Liu B, Song B, Su Y. The clinical dynamic changes of macrophage phenotype and function in different stages of human wound healing and hypertrophic scar formation. International Wound Journal 2019; 16: 360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilkinson HN, Roberts ER, Stafford AR, Banyard KL, Matteucci P, Mace KA, Hardman MJ Tissue Iron Promotes Wound Repair via M2 Macrophage Polarization and the Chemokine (C-C Motif) Ligands 17 and 22. The American journal of pathology 2019; 189: 2196–2208. [DOI] [PubMed] [Google Scholar]
- 63.Driscoll WS, Vaisar T, Tang J, Wilson CL, Raines EW Macrophage ADAM17 deficiency augments CD36-dependent apoptotic cell uptake and the linked anti-inflammatory phenotype. Circulation research 2013; 113: 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee SJ, Evers S, Roeder D, Parlow AF, Risteli J, Risteli L, Lee YC, Feizi T, Langen H, Nussenzweig MC Mannose Receptor-Mediated Regulation of Serum Glycoprotein Homeostasis. Science 2002; 295: 1898. [DOI] [PubMed] [Google Scholar]
- 65.Sallusto F, Cella M, Danieli C, Lanzavecchia A. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. The Journal of experimental medicine 1995; 182: 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sironi M, Martinez FO, D’Ambrosio D, Gattorno M, Polentarutti N, Locati M, Gregorio A, Iellem A, Cassatella MA, Van Damme J, Sozzani S, Martini A, Sinigaglia F, Vecchi A, Mantovani A. Differential regulation of chemokine production by Fcγ receptor engagement in human monocytes: association of CCL1 with a distinct form of M2 monocyte activation (M2b, Type 2). Journal of Leukocyte Biology 2006; 80: 342–349. [DOI] [PubMed] [Google Scholar]
- 67.Nishiguchi T, Ito I, Lee JO, Suzuki S, Suzuki F, Kobayashi M. Macrophage polarization and MRSA infection in burned mice. Immunology and cell biology 2017; 95: 198–206. [DOI] [PubMed] [Google Scholar]
- 68.Novak ML and Koh TJ Macrophage phenotypes during tissue repair. Journal of leukocyte biology 2013; 93: 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Philippidis P, Mason J, Evans B, Nadra I, Taylor K, Haskard D, Landis R. Hemoglobin scavenger receptor CD163 mediates interleukin-10 release and heme oxygenase-1 synthesis: antiinflammatory monocyte-macrophage responses in vitro, in resolving skin blisters in vivo, and after cardiopulmonary bypass surgery. Circulation research 2004; 94: 119–126. [DOI] [PubMed] [Google Scholar]
- 70.Jenner W, Motwani M, Veighey K, Newson J, Audzevich T, Nicolaou A, Murphy S, MacAllister R, Gilroy DW Characterisation of Leukocytes in a Human Skin Blister Model of Acute Inflammation and Resolution. PloS one 2014; 9: e89375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karna E, Szoka L, Huynh TYL, Palka JA Proline-dependent regulation of collagen metabolism. Cellular and Molecular Life Sciences 2020; 77: 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Litwiniuk M, Krejner A, Speyrer MS, Gauto AR, Grzela T. Hyaluronic acid in inflammation and tissue regeneration. Wounds 2016; 28: 78–88. [PubMed] [Google Scholar]
- 73.Galdiero MR, Garlanda C, Jaillon S, Marone G, Mantovani A. Tumor associated macrophages and neutrophils in tumor progression. Journal of cellular physiology 2013; 228: 1404–1412. [DOI] [PubMed] [Google Scholar]
- 74.Wenes M, Shang M, Di Matteo M, Goveia, Martín-Pérez R, Serneels Prenen H, Ghesquière B, Carmeliet P, Mazzone M Macrophage Metabolism Controls Tumor Blood Vessel Morphogenesis and Metastasis. Cell Metabolism 2016; 24: 701–715. [DOI] [PubMed] [Google Scholar]
- 75.Yan D, Kowal J, Akkari L, Schuhmacher AJ, Huse JT, West BL, Joyce JA Inhibition of colony stimulating factor-1 receptor abrogates microenvironment-mediated therapeutic resistance in gliomas. Oncogene 2017; 36: 6049–6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arts RJ, Plantinga TS, Tuit S, Ulas T, Heinhuis B, Tesselaar M, Sloot Y, Adema GJ, Joosten LA, Smit JW Transcriptional and metabolic reprogramming induce an inflammatory phenotype in non-medullary thyroid carcinoma-induced macrophages. Oncoimmunology 2016; 5: e1229725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu D, Chang C, Lu N, Wang X, Lu Q, Ren X, Ren P, Zhao D, Wang L, Zhu Y. Comprehensive proteomics analysis reveals metabolic reprogramming of tumor-associated macrophages stimulated by the tumor microenvironment. Journal of Proteome Research 2017; 16: 288–297. [DOI] [PubMed] [Google Scholar]
- 78.Gabriel VA, McClellan EA, Scheuermann RH Response of human skin to esthetic scarification. Burns 2014; 40: 1338–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van den Bossche J, Baardman J, Otto NA, van der Velden S, Neele AE, van den Berg SM, Luque-Martin R, Chen H-J, Boshuizen MC, Ahmed M. Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell reports 2016; 17: 684–696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.