Figure 11. Selection of a bad highly variable gene set leads to suboptimal performance and obscures biological signal.
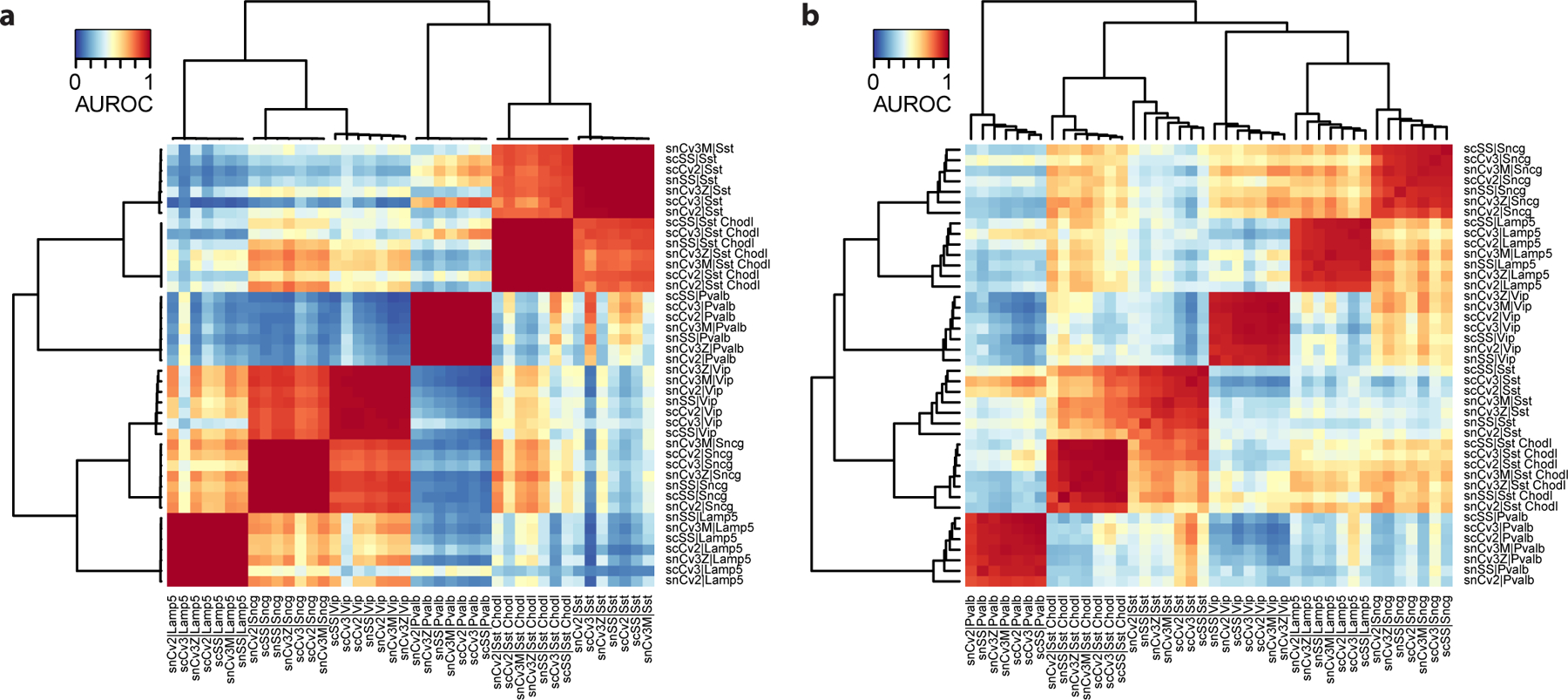
a Anticipated result: AUROC heatmap based on a set of highly variable genes selected by MetaNeighbor. The heatmap has clear replicating clusters (dark red squares) and known secondary biological relationships (e.g., similarity of CGE-derived interneurons Vip, Sncg and Lamp5). b Possible issue: AUROC heatmap based on a set of random genes (same number of genes as the correctly selected highly variable gene set in a). Replicability patterns become weaker: lower performance, gradients within replicating cell types, weaker secondary relationships.
