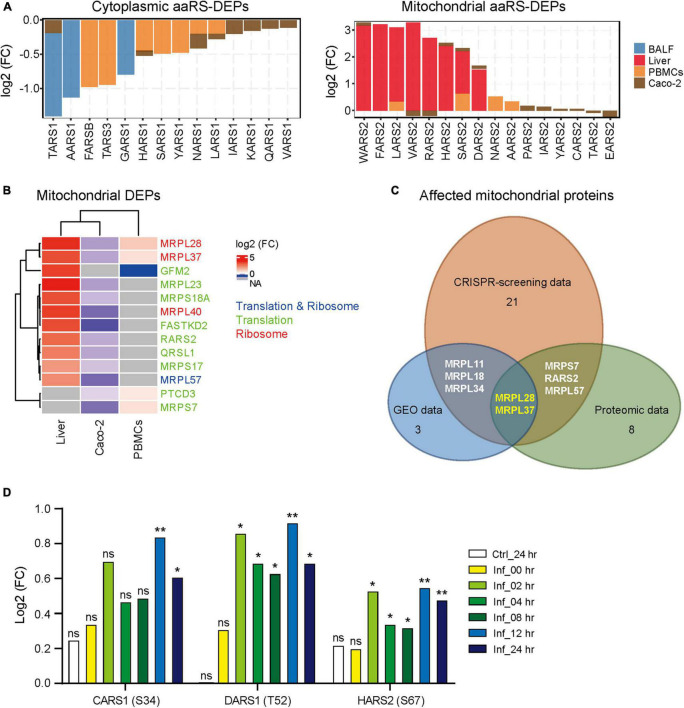
FIGURE 3.
Protein profile and phosphorylated modification of aminoacyl-tRNA synthetases (aaRSs) upon SARS-CoV-2 infection. (A) Protein levels of aaRS-DEPs (with the cyto-aaRSs on the left and the mt-aaRSs on the right) in BALF, liver cells and PBMCs from coronavirus disease 2019 (COVID-19) patients, as well as in Caco-2 cells infected with SARS-CoV-2. (B) Heatmap of mitochondrial translation- and ribosome-related DEPs from different samples. The fold change in protein expression is indicated by color intensity, with red representing the upregulated DEPs and blue representing the downregulated DEPs. The DEPs include translation- and ribosome-related proteins (blue), translation-related proteins (green), and ribosome-related proteins (red). (C) Wayne diagram of the affected mitochondrial translation- and ribosome-related genes and proteins across CRISPR-screening, gene expression omnibus (GEO), and proteomic data. Genes with significant Z scores upon SARS-CoV-2 infection, mitochondrial DEGs and DEPs were included for analysis. The remaining number of the affected mitochondrial proteins in each platform is indicated except for the overlapping ones. (D) Phosphorylation of aaRSs (phosphorylated site) at different indicated time. Ctrl, control; Inf, infection; hr, hour; ns, not significant; *p < 0.05; **p < 0.01 (vs. mock-infected cells at 0 h).