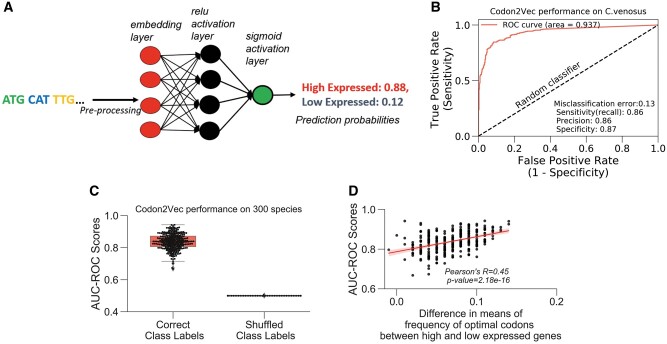
Fig. 6.
Neural network uses codons as features to predict gene expression. (A) Schema of Codon2Vec. Codon2vec is a fully connected multilayer neural network that uses an embedding layer to transform codons in the input coding sequences to a real-valued vector. The final output of the model is a vector of probabilities for each gene expression class (i.e., “high” or “low”). Detailed description in Materials and Methods. (B) Codon2Vec’s performance on a single species. Model performance is evaluated on the hold-out test sets based on the AUC-ROC. An AUC score of 0.5 (dashed line) represents random predictions. (C) Model generalizability: Codon2Vec achieves high predictive performance on 300 different species. However, shuffling of ground truth labels independent of input sequences ablated Codon2Vec’s ability to learn meaningful associations. (D) Codon2Vec’s prediction accuracy (AUC-ROC) positively and significantly correlates with differential usage of optimal codons between high and low expressed genes. The frequency of optimal codons (FOP) is another standard metric for codon usage bias (Ikemura 1982; Materials and Methods).