Extended Data Fig. 5 |. Comparisons between CaSR cryo-EM structures and previous family C GPCR structures.
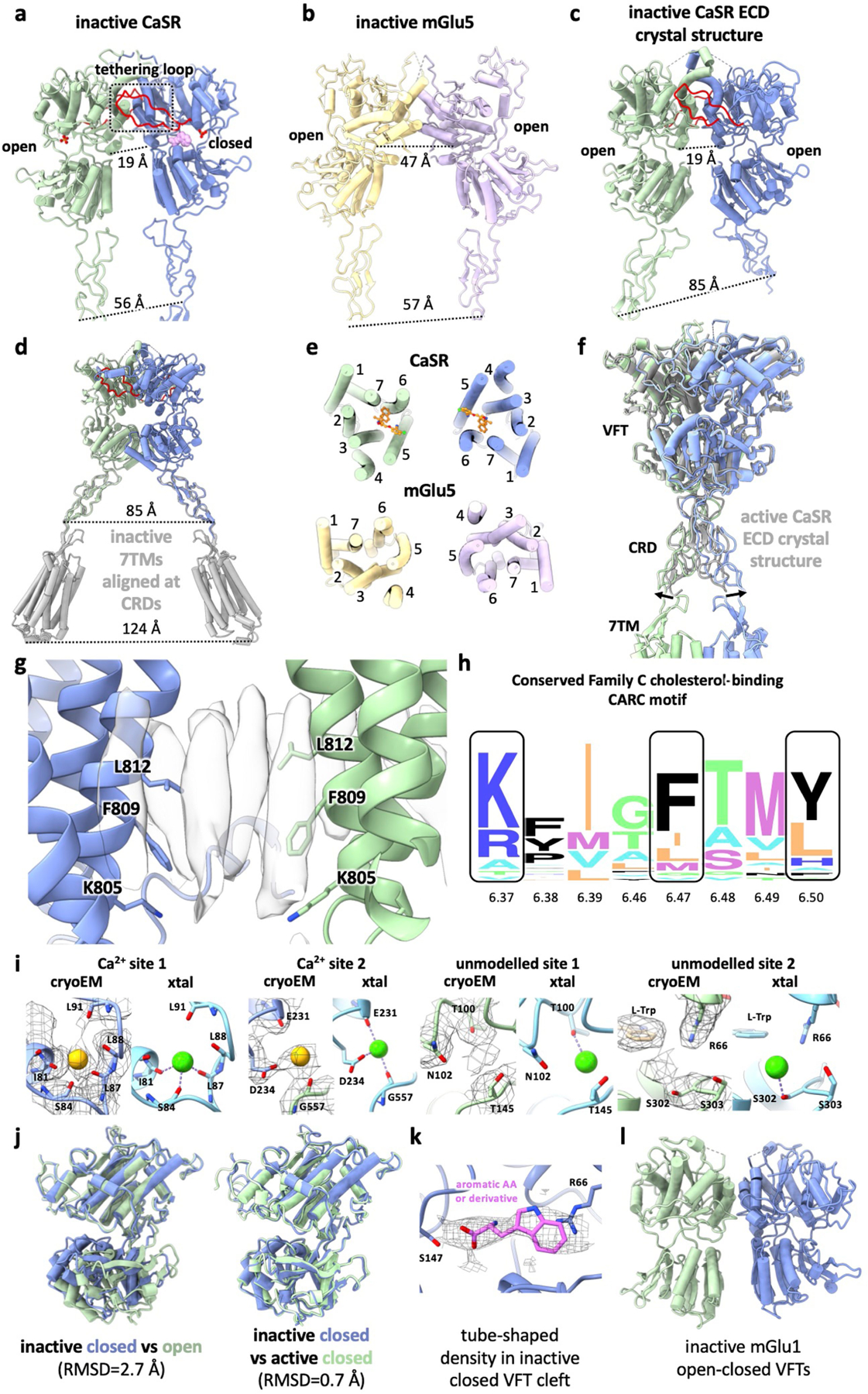
a, ECD from inactive-state CaSR cryo-EM structure with the loop tethering the opposing LB1 colored in red. b, ECD from inactive-state mGlu5 cryo-EM structure8 (PDB: 6N51). c, inactive-state CaSR ECD crystal structure6 (PDE: 5K5T). d, 7TMs from inactive-state CaSR cryo-EM structure (grey) superposed onto inactive CaSR ECD crystal structure based on CRD alignment. e, Comparison of inactive 7TMs orientations between CaSR and mGlu5. f, Active-state CaSR cryo-EM structure aligned with the active CaSR ECD crystal structure6 (grey, PDB: 5K5S) illustrating the difference in CRD orientations. g, Elongated densities observed at the TM6-TM6 interface in cryo-EM maps of active-state CaSR shown with TM6 residues forming the cholesterol-binding CARC motif. h, Sequence alignment logo showing the conservation of CARC motif residues among family C GPCRs (generated using alignment from GPCRdb43 with WebLogo44). i, Comparison of Ca2+ sites in active-state cryo-EM structures with Ca2+ sites in active CaSR ECD crystal structure (PDB: 5K5S)6. j, Alignment of the closed VFT protomer (dark blue) observed in our inactive-state CaSR structure with either the inactive open protomer (left, dark green) or the active closed protomer (right, light green). k, The tube shaped density observed in closed inactive CaSR VFT with L-Trp docked in. l, Crystal structure of open-closed mGlu1 VFTs13 (PDB: 1EWV).
