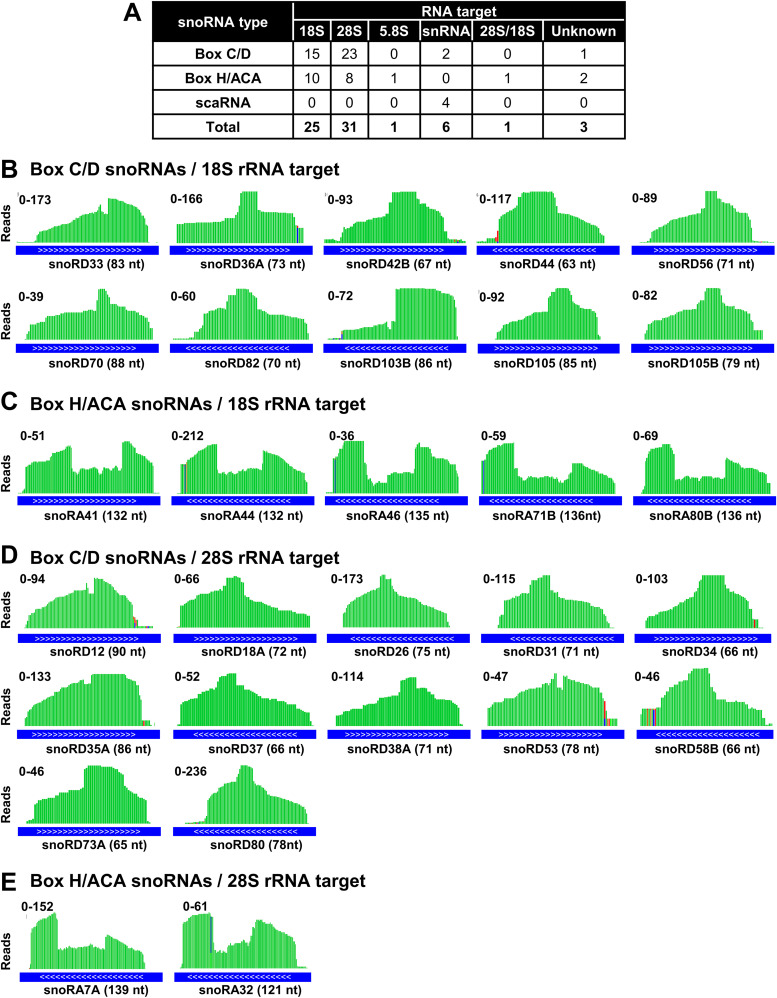
FIG 5.
ORF57-associated snoRNAs target rRNAs. (A) A table showing the type and number of predicted RNA targets from snoRNAs associated with ORF57. snoRNA type and target RNA were obtained from the snOPY database http://snoopy.med.miyazaki-u.ac.jp/. (B and C) IGV visualization of read distribution of selective box C/D (B) and H/ACA (C) snoRNAs guiding 18S rRNA modifications which bind to ORF57. The snoRNAs shown were identified by ORF57 CLIP-seq with a P value of ≤1 × 10−15 and number of reads of ≥70 and also validated by ORF57 RIP-snoRNA array analyses. ENSEMBL transcript annotation tracks (blue lines) shown at the bottom indicate the orientation (white arrows 5′ to 3′) and size in nucleotides of each snoRNA. Values on the y axis represent the scale of ORF57 CLIP-seq read counts. (D and E) IGV visualization of read distribution of selective box C/D (D) and H/ACA (E) snoRNAs guiding 28S rRNA modification which bind to ORF57. See other details described for panels B and C.