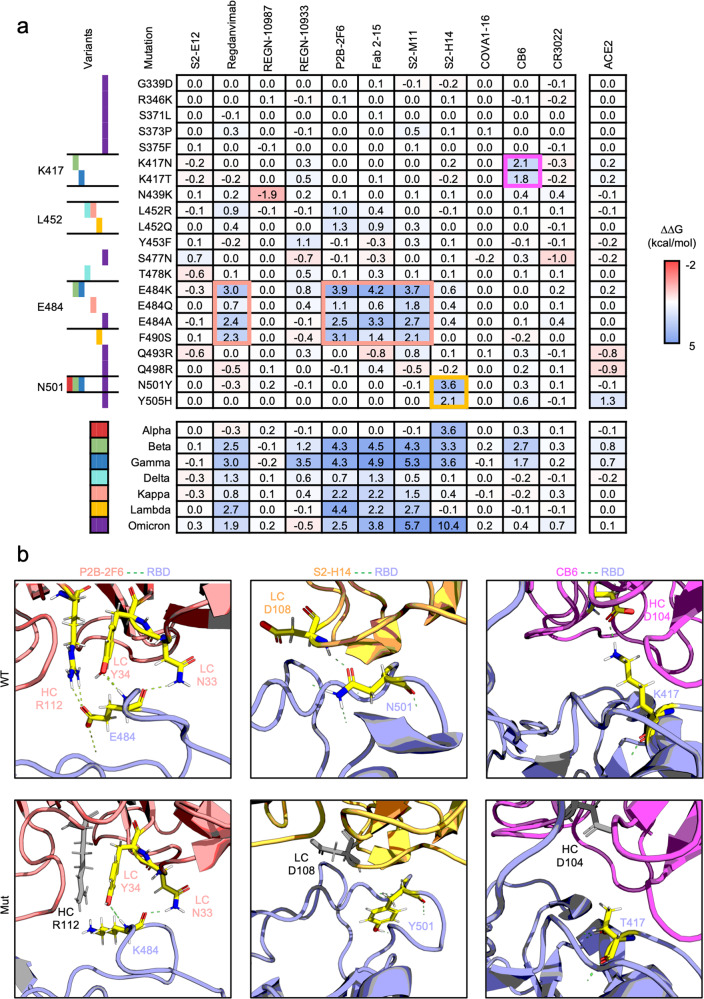
Fig. 1.
Computational interface analysis of SARS-CoV-2 spike protein variants by FlexddG. a Interfacial free energy (∆∆G) heatmap for complexes of neutralizing antibodies or ACE2 with RBD variants containing the indicated mutations. The ∆∆G values are shown in the table. b Representation of the structural interface of the antibodies P2B-2F6, S2-H14, and CB6 with wild-type or mutated RBD. The interacting residues are shown as sticks and colored by atom (carbon: yellow, hydrogen: white, nitrogen: blue, oxygen: red). The hydrogen bonds are displayed as green dashed lines. Residues in antibodies that lost hydrogen bonds to the RBD due to the indicated mutations are gray. The shown ΔΔG of each antibody or receptor in contact with the indicated mutation of the RBD is the average value from 35 output structures generated by flexddG