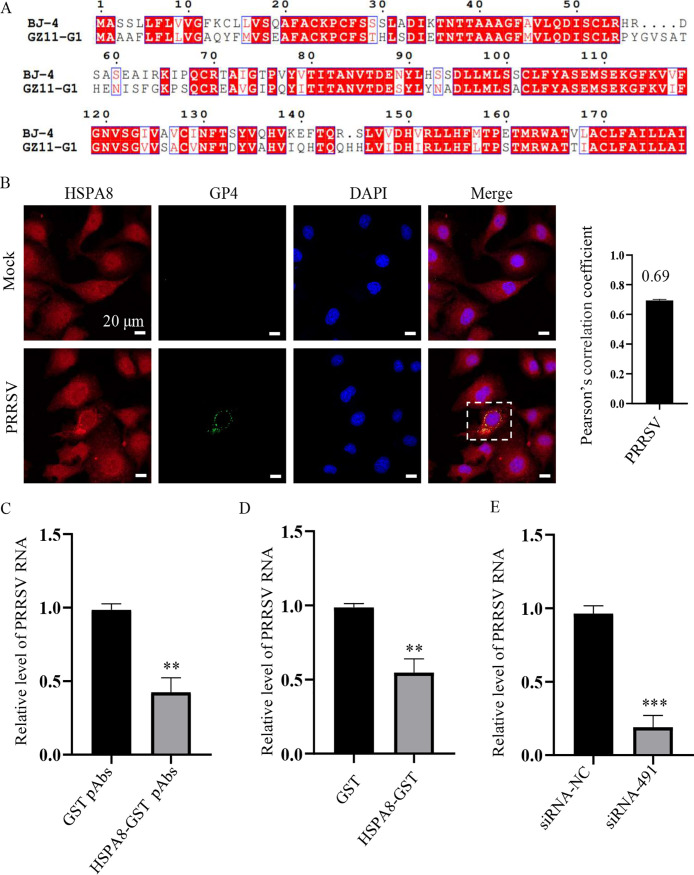
FIG 8.
HSPA8 is important for PRRSV-1 attachment and internalization. (A) Amino acid sequence alignment of GP4 from PRRSV-1 strain GZ11-G1 (GenBank: KF001144) and PRRSV-2 strain BJ-4 (GenBank: AF331831). ClustalW and ENDscript/ESPript software were used for comparative analysis. (B) PRRSV-1 GZ11-G1 co-localized with HSPA8 in MARC-145 cells during early infection. MARC-145 cells were infected with PRRSV-1 GZ11-G1 (1 MOI) at 37°C for 24 h. The cells were washed with PBS, fixed with 4% PFA, permeabilized with 0.1% Triton X-100, and stained with anti-HSPA8 pAbs (catalog no. 10654-1-AP; red) and anti-PRRSV GP4 pAbs (green). The cell nuclei were stained with DAPI (blue). Images were acquired on the Zeiss confocal microscope. The Pearson’s overlap coefficient was analyzed. The mock-infected cells were used as control. Scale bars, 10 μm. (C) HSPA8 pAbs inhibited PRRSV-1 GZ11-G1 infection. MARC-145 cells were incubated with HSPA8-GST pAbs and GST pAbs at 1:16 dilution in DMEM at 37°C for 1 h. Then the cells were washed with PBS and inoculated with PRRSV-1 GZ11-G1 (1 MOI) at 4°C for 1 h. After three washes, the cells were again incubated with DMEM containing the corresponding antibodies for 24 h. PRRSV RNA abundance was determined by RT-qPCR. Data represent means ± SD from three independent experiments. **, P < 0.01. (D) Soluble HSPA8 protein inhibited PRRSV-1 infection. PRRSV-1 GZ11-G1 at 1 MOI was incubated with HSPA8-GST or GST protein at the final concentration of 2 μM at 37°C for 1 h and then inoculated in MARC-145 cells and harvested at 24 h. PRRSV RNA abundance was determined by RT-qPCR. Data represent means ± SD from three independent experiments. **, P < 0.01. (E) HSPA8 knockdown decreased PRRSV-1 GZ11-G1 infection. MARC-145 cells were transfected with siRNA-491 and siRNA-NC for 36 h, and then infected with PRRSV-1 GZ11-G1 at 1 MOI for 24 h. PRRSV RNA abundance was determined by RT-qPCR. Data represent means ± SD from three independent experiments. ***, P < 0.001.