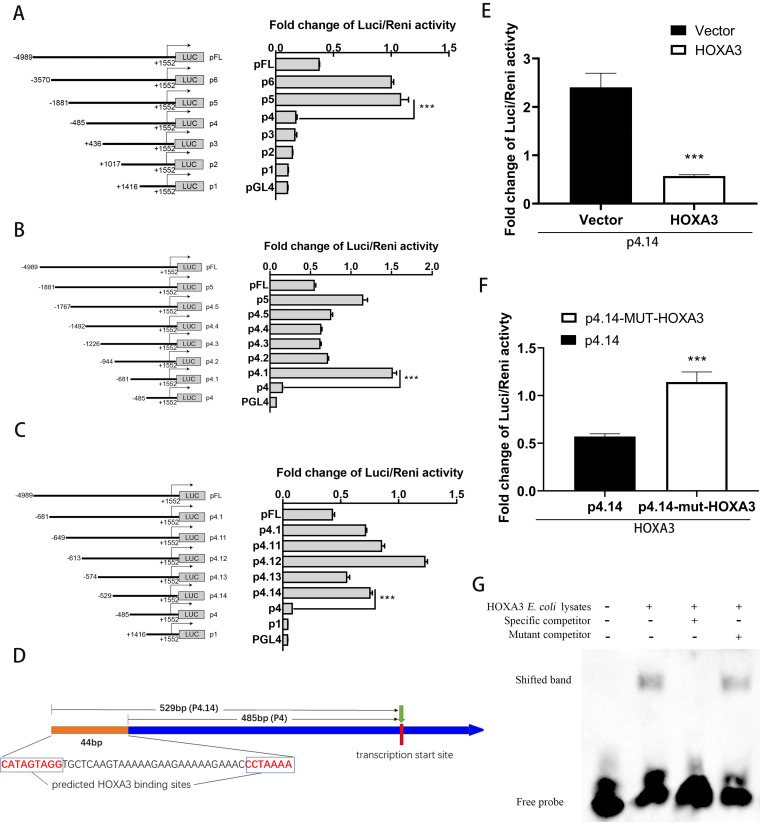
FIG 1.
HOXA3 is a new transcription factor of the HO-1 gene. (A to C) A series of truncation plasmids of pGL4 luciferase reporter assays containing a variety of fragments of the HO-1 gene promoter region was constructed. HEK293T cells were transfected with plasmids for 24 h, and cells were lysed to detect different truncated promoter regions of HO-1 gene luciferase activity. (D) Schematic illustration of HOXA3 binding site in the promoter region of the HO-1 gene. (E) HEK293T cells were cotransfected with pGL4-p4.14 and pCAGEN-HOXA3-myc for 36 h; the empty vector of pCAGEN-myc was used as the control. The luciferase activity was measured with a dual-luciferase assay. (F) HEK293T cells were cotransfected with pGL4-p4.14-mut-HOXA3 and pCAGEN-myc-HOXA3 for 36 h; the plasmid of pGL4-p4.14 was used as the control. The luciferase activity was measured with a dual-luciferase assay. (G) EMSA. Lane 1, biotin‐labeled WT probe. Lane 2, biotin‐labeled WT probe +HOXA3 E. coli lysates. Lane 3, biotin‐labeled WT probe + HOXA3 E. coli lysates + 100‐fold molar excess of biotin‐unlabeled specific competitor probe. Lane 4, biotin‐labeled WT probe + HOXA3 E. coli lysates + 100‐fold molar excess of biotin‐unlabeled mutant competitor probe. The details were described in the Materials and Methods. Results are expressed as means ± SD of three independent replicates. Statistically significant values were denoted as follows: ***, P < 0.001.