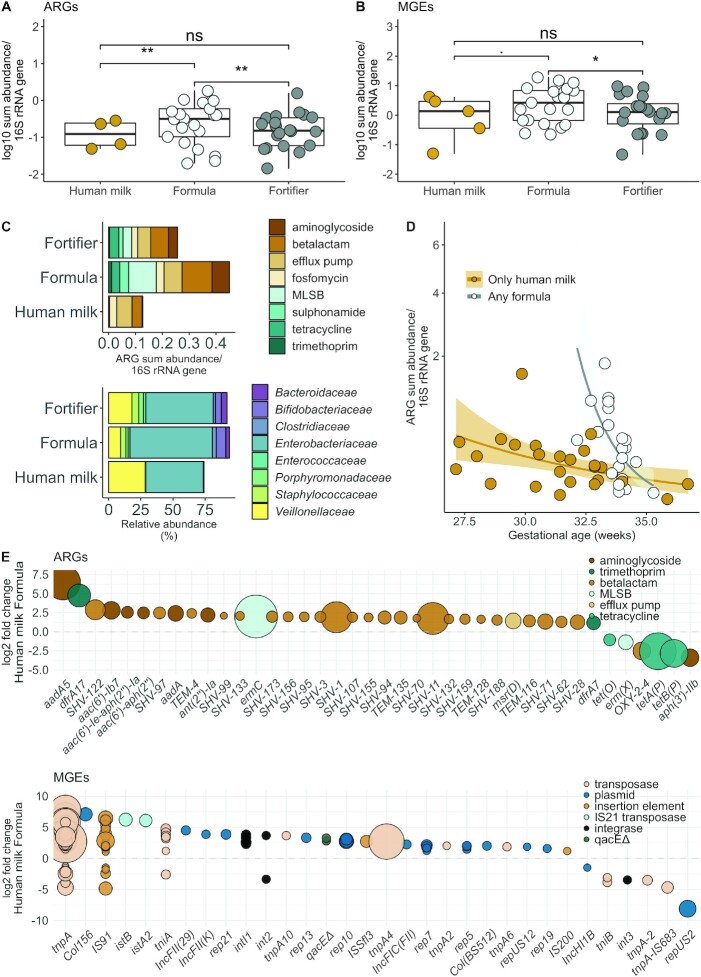
FIGURE 3.
Effects of diet and gestational age on ARGs and MGE abundances in the preterm infant cohort. (A) Relative ARG and (B) relative MGE sum abundances by a diet containing either only human milk, any formula, or human milk and fortifier (n = 46). Infants fed any formula had higher ARG abundances (normalized to 16S rRNA counts) than infants fed only breast milk or fed milk supplemented with fortifier [gamma-distributed GLM adjusted with gestational age and 16S rRNA counts, fold changes 4.3 (95% CI: 1.61–11.56) and 3.6 (95% CI: 1.61–8.9), respectively; adjusted P < 0.01]. MGEs were significantly more abundant (normalized to 16S rRNA gene) in the infants fed any formula than in infants fed exclusively human milk (gamma-distributed GLM, fold change 2.07 (95% CI: 1.08–3.99; P < 0.05). (A and B) The y axis denotes log10 transformed ARG or MGE copies normalized by 16S rRNA gene copied counted from shotgun sequence data. Significance levels are denoted as follows: ** = 0.001–0.01; * = 0.01–0.05; . = 0.05–0.1; ns = 0.1–1. The boxplot hinges represent 25% and 75% percentiles and centerline the median. Notches are calculated with the formula median ± 1.58 × IQR/sqrt (n). (C) Differences between most abundant ARG classes and bacterial families. The x axis represents ARG copies normalized to 16S rRNA gene counts and percentage of bacterial family. (D) Effect of gestational age and formula on relative ARG abundances. The x axis represents gestational age in weeks. The y axis has been square root transformed. (E) Differentially abundant ARGs and MGEs using DESeq2 and an adjusted P value cutoff of 0.05 for the reported genes. Genes with positive values are more abundant in formula-fed infants. The larger the point size, the more abundant the gene is in the samples. Abbreviations: ARG, antibiotic resistance gene; GLM, generalized linear mode; MGE, mobile genetic element; MLSB, macrolide-lincosamide-streptogramin B; rRNA, ribosomal RNA.