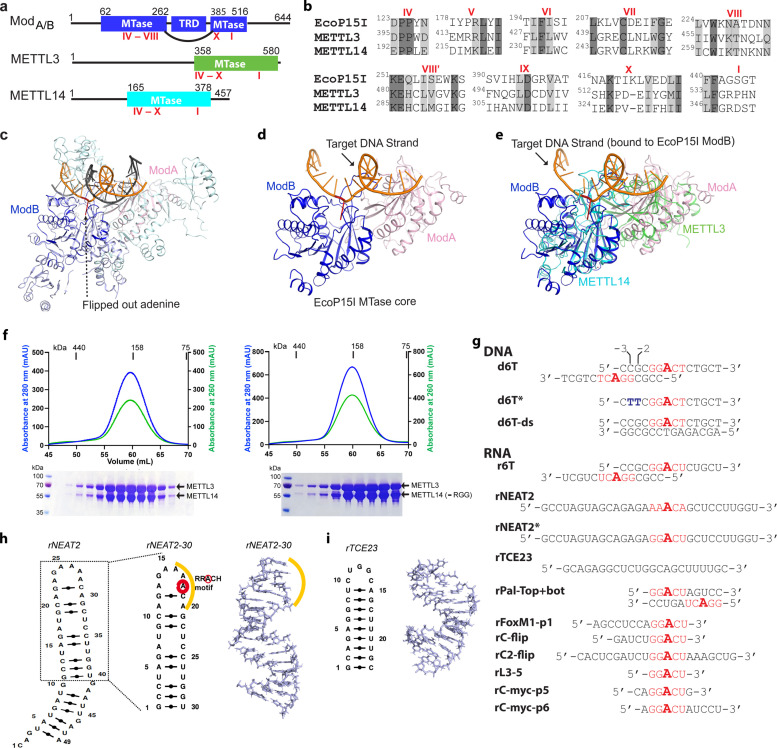
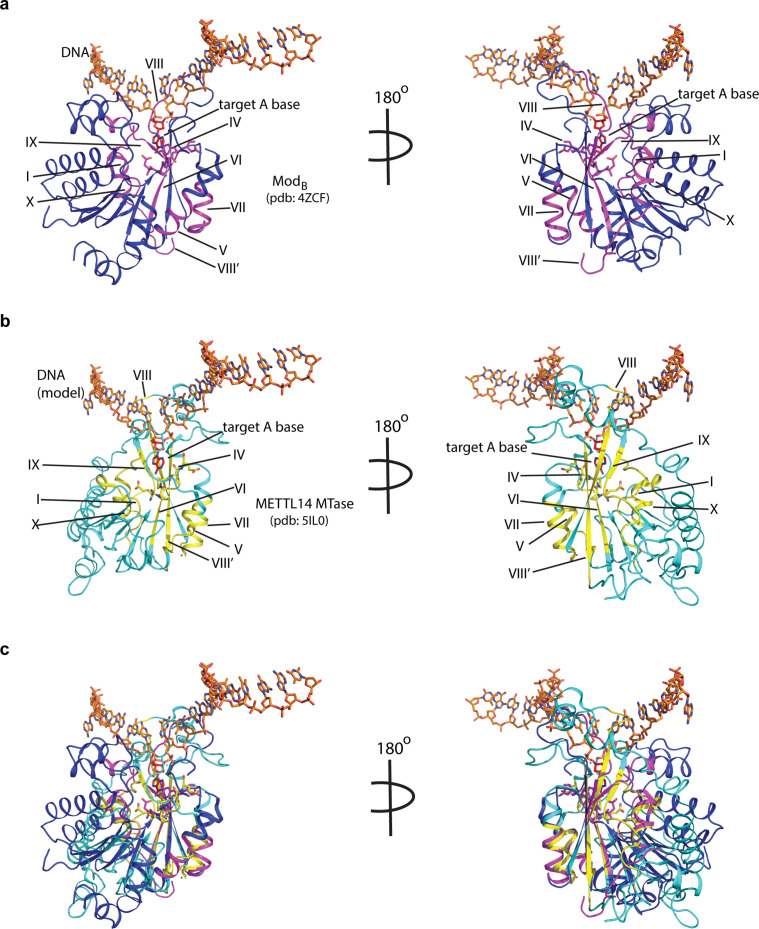
Figure 1. Structural similarity, purification of methyltransferases, and substrate designing.
(a–b) Domain architecture of Mod subunit of EcoP15I, human methyltransferase like-3 (METTL3), and human METTL14 methyltransferases (MTases). All three members belong to the β-class of SAM-dependent MTases and exhibit a sequential arrangement of motifs (IV-X followed by motif I) (Bujnicki et al., 2002; Woodcock et al., 2020; Malone et al., 1995). Motif IV (D/EPPY/W/L) and I are associated with the recognition of target adenine base (Gupta et al., 2015) and co-factor (SAM) binding (Wang et al., 2016b; Sledz and Jinek, 2016; Wang et al., 2016a), respectively. (c) Crystal structure of EcoP15I-DNA complex (PDB ID: 4ZCF) (Gupta et al., 2015): ModA MTase (cyan), ModB MTase (blue), non-methylating DNA strand (gray), target (methylating) DNA strand (orange), flipped adenine base (red stick). The methyltransferase cores of ModB and ModA are shown in dark blue and light pink, respectively. The Res subunit of EcoP15I was omitted for clarity. (d) MTase domains of two Mod subunits (ModA/B) of EcoP15I with target DNA strand. The regions flanking the methyltransferase core (e.g., CTD and TRD) are omitted for clarity. Only the region encompassing the MTase core (aa 90–132, 169–261, and 385–511) of EcoP15I Mod was selected for the alignment with the methyltransferase core of METTL3 (aa 358–580) and METTL14 (aa 165–378). (e) An overlay of MTase domains of EcoP15I and METTL14 (PDB ID: 5IL0) shows structural similarity within MTase folds (rmsd = 2.55 Å over 278 Cα atoms). RNA strand here was modeled based on the respective methylating strand in the EcoP15I structure.(f) Chromatogram of final size exclusion chromatography (SEC) step of purification showing METTL3-METTL14 complexes (left, full-length; right, METL3-METTL14[-RGG]) co-eluted as single homogenous species. Blue, absorbance at 280 nm; green, absorbance at 260 nm (A260). Coomassie stained gels (lower panels) confirm high purity of METTL3-METTL14 proteins in the SEC peak fractions. (g) Sequence of DNA and RNA oligonucleotides used in this study. All oligos have a covalently attached 5’-fluorescein (not shown). (h) Secondary structure of rNEAT2 and its 3D model as predicted by MC-SYM (Parisien and Major, 2008). rNEAT2 harbors a potential DRACH motif (yellow line). (i) Secondary structure of rTCE23 RNA and its solution NMR structure (PDB ID: 2ES5) (Oberstrass et al., 2006).