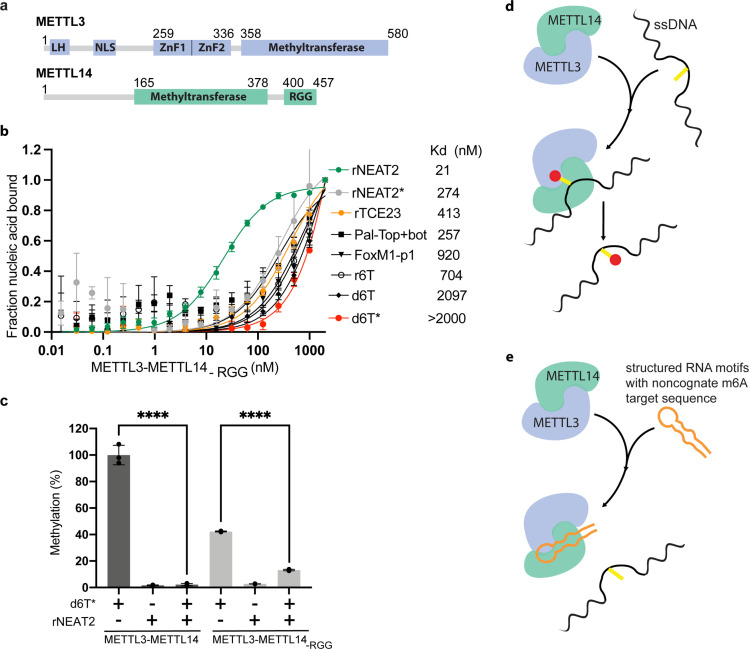
Figure 3. Role of RGG motifs and model of RNA-mediated regulation of methyltransferase activity.
(a) Domain architecture of methyltransferase like-3 (METTL3) and METTL14. LH, leader helix; NLS, nuclear localization signal; ZnF1/2, zinc-finger domain 1/2; RGG, arginine-glycine rich repeats motif. (b) FP-based-binding assay for DNA and RNA oligos showing the highest affinity of METTL3-METTL14(-RGG) for rNEAT2 RNA (green) and lowest affinity for d6T* (red). Equilibrium dissociation constants (Kd) for each oligo are shown. The data were fit into one site-specific binding model (Y = Bmax*X/(Kd+ X)). See Materials and methods section and source data for details. (c) Relative methyltransferase activity of full-length METTL3-METTL14 and the truncated enzyme devoid of the RGG motif in METTL14 ([METTL3-METTL14(-RGG)]) in the presence of d6T*, rNEAT2, or an equimolar mixture of these two oligos, as measured by radiometric assay. Results presented are the average of three independent experiments (n = 3) with one standard deviation (s.d.) for each oligonucleotide (shown as error bars). The results of two groups were analyzed and compared using two-tailed Student’s unpaired t-test (p-value < 0.0001). Details about Student’s t-test are provided in the Source Data file. (d, e) Proposed models showing that the METTL3-METTL14 complex can methylate a target adenine (yellow) in a single-stranded DNA region (black). Structured motifs present in ncRNA/mRNAs (orange) can block the methyltransferase activity by a shape-dependent binding of these RNAs to METTL3-METTL14.