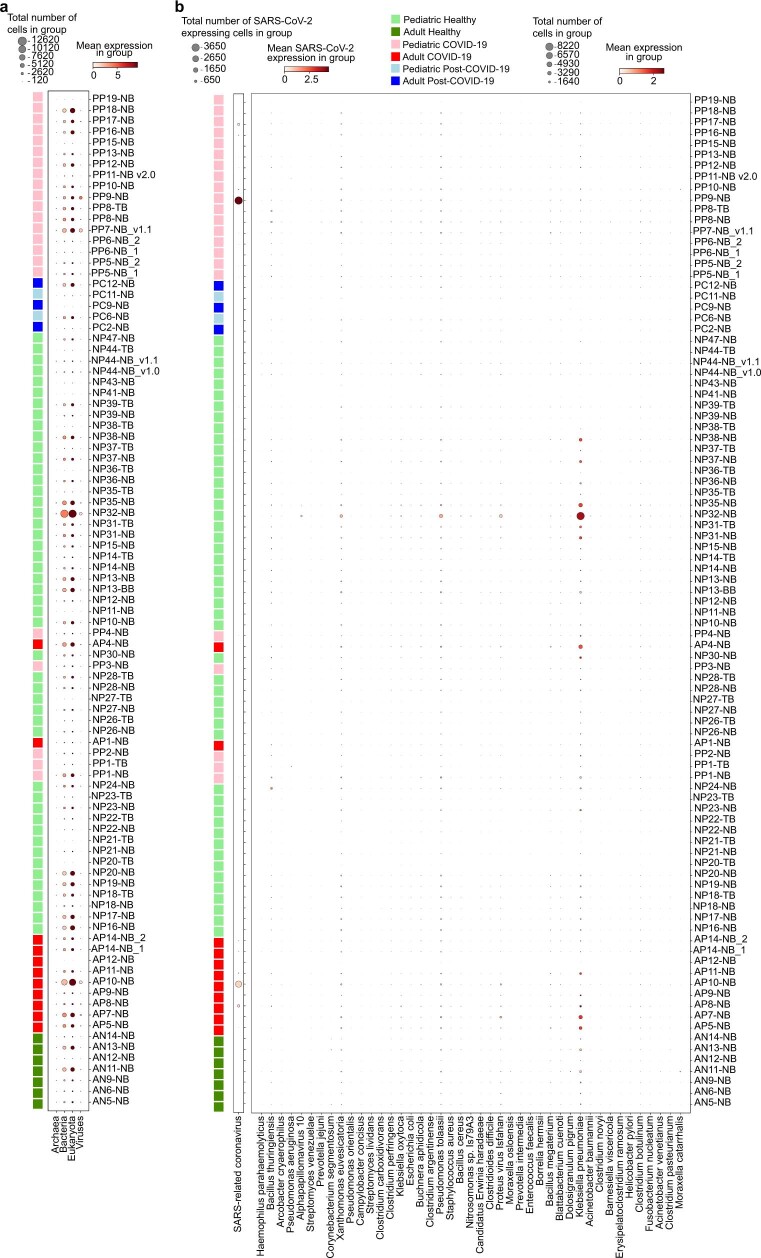
Extended Data Fig. 9. Metagenomic analysis of patient sample reads that were not mapped to the human genome.
(a) Dotplot showing the amount of cells that harbour reads aligned to archaea, bacteria, eukaryota (including human reads that initially did not align to the human transcriptome by STARsolo) and viruses. (b) Dotplot showing the amount of cells that harbour reads to a selection of disease-relevant bacteria and viruses. Apart from SARS-CoV-2 and non-specific signal found in most samples, we did not detect any pathogens that were highly abundant in samples of interest.