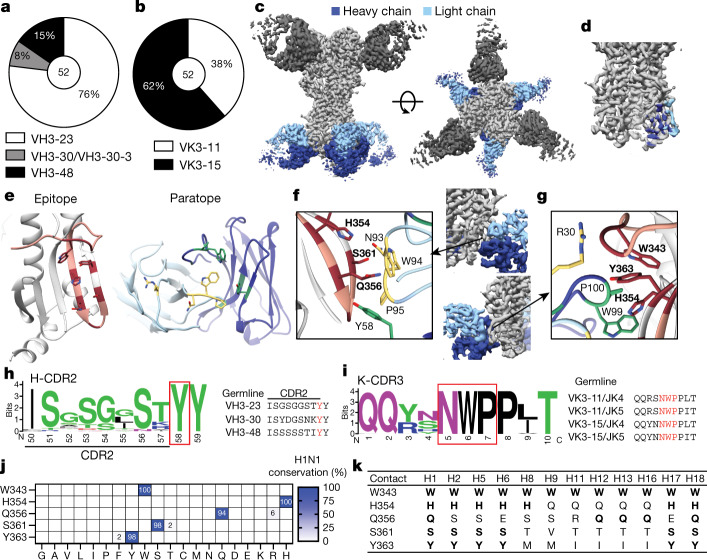
Fig. 2. Anchor-targeting mAbs bind to the HA fusion peptide via public binding motifs.
a, b, VH (a) and VK (b) gene usage by anchor-binding mAbs. The number in the centre of the pie graphs indicates the number of mAbs analysed. c, Cryo-EM structure of anchor-targeting 222-1C06 (blue) and lateral patch-targeting 045-09 2B05 (dark grey; see Methods) binding to A/California/7/2009 (E376K) HA (light grey). d, Heavy chain and light chain footprint of 222-1C06 binding to HA based on the cryo-EM structure. e, HA epitope contact residues (maroon) and heavy chain (green) and light chain (yellow) antibody contact residues of the 222-1C06 paratope. Peach-highlighted amino acids represent the fusion peptide of HA2. f, K-CDR3 NWP and H-CDR2 Y58 motifs of 222-1C06. Bold residues are HA residues. g, Major contacts of 222-1C06 K-CDR1 and H-CDR3 (normal typeface) binding to HA (bold residues). h, i, Weblogo plot and germline sequence of Y58 following the H-CDR2 motif (h) and the K-CDR3 NWP motif (i). j, k, Amino acid conservation of s contact residues across human, swine and avian H1 viruses (j) and group 1 influenza A viruses (k). Bold residues are contacts conserved with A/California/7/2009 H1N1 (k). The strain information used for conservation models in j and k are in Supplementary Tables 4, 5, respectively. See also Extended Data Fig. 4.