Figure 3.
DOT1L inhibition leads to transcriptional changes not observed in MEFs or ESCs
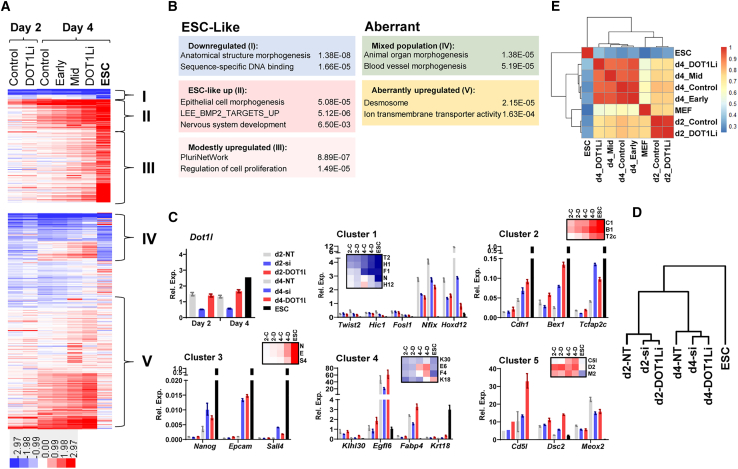
(A) k-means clustering of DOT1Li-DE genes organized based on transcriptional change in ESCs relative to MEFs. Heatmap intensity indicates log2 FC TPM relative to MEF for each sample.
(B) Representative gene ontology categories for each cluster. The most enriched categories containing unique sets of genes are displayed (HOMER).
(C) qPCR validation of select genes from every cluster in non-targeting control (NT) treated with control DMSO, siDot1l (si), and DOT1Li. Normalized expression in MEFs was set to 1 for Dot1l and clusters 1, 4, and 5. Normalized expression in ESCs was set to 1 for clusters 2 and 3. All samples shown are from day 2 (left) to day 4 (right). Insets: zoomed-in RNA-seq heatmaps of assessed genes in control (C), DOT1Li (D), and ESCs at day 2 (2) and day 4 (4). Genes are abbreviated as first initial and number.
(D) Hierarchical clustering of relative expression of tested genes (C).
(E) Spearman correlation of DOT1Li-DE gene TPM across all samples.
See also Figure S3.