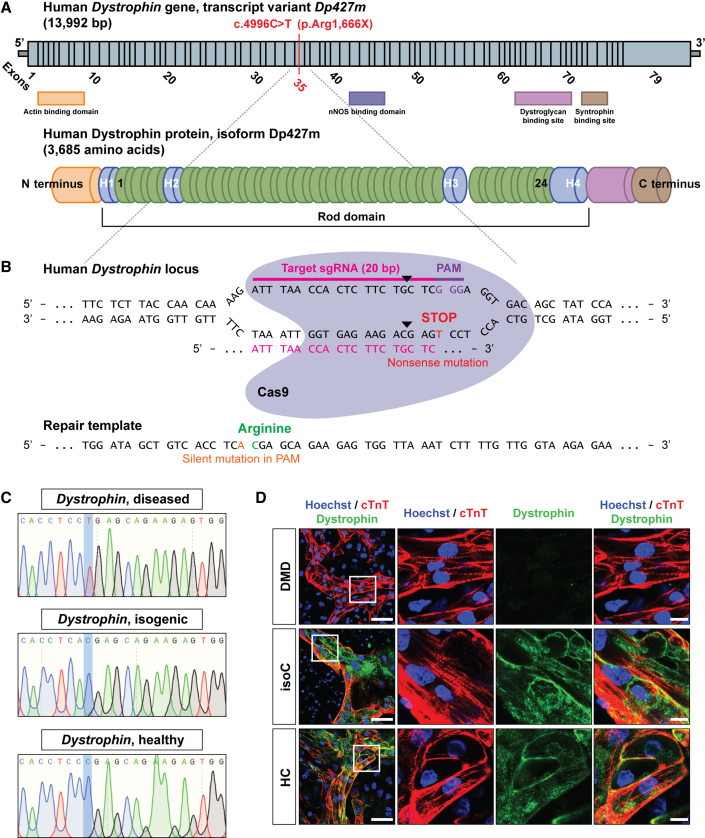
Figure 1.
DMD hiPSC CRISPR-Cas9 gene editing of a nonsense mutation in exon 35 (c.4,996C>T; (p.Arg1,666X)) of the Dystrophin gene
(A) Schematic representation of the human Dystrophin gene sequence (top, transcript variant Dp427m) and the encoded Dystrophin protein (bottom, isoform Dp427m). The genetic point mutation is located in exon 35 of the Dystrophin gene, resulting in a premature stop codon.
(B) The 20 nt sgRNA (ATTTAACCACTCTTCTGCTC) to induce the Cas9-mediated DSB (indicated as black triangles). The donor repair template containing the genetic correction of the nonsense mutation in the Dystrophin gene is also shown.
(C) DNA sequencing of the mutated region of interest of Dystrophin before (DMD diseased) and after (DMD isogenic) CRISPR-Cas9 gene editing.
(D) Immunofluorescent staining showing the expression of Dystrophin protein levels (green) in differentiated DMD hiPSC-CMs (cTnT, red and Hoechst, blue) after CRISPR-Cas9-mediated correction. Scale bar: 50 μm. White boxes with corresponding insets are at a higher magnification. Scale bar: 10 μm.
See also Figures S1F, S1G, and S2A–S2D.