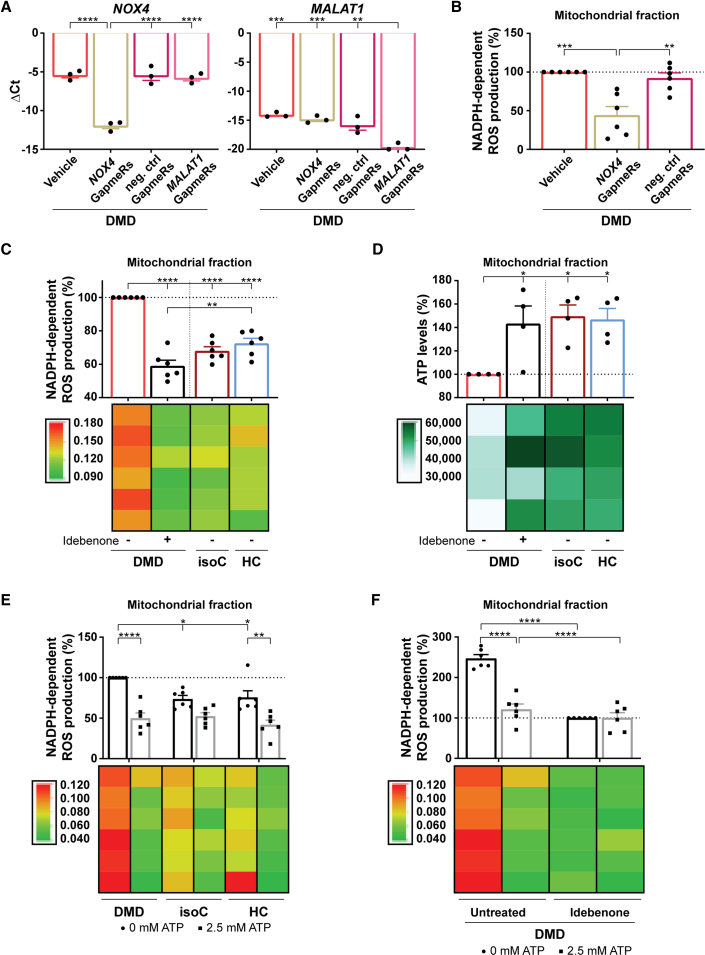
Figure 6.
Idebenone could counteract the oxidative stress in DMD hiPSC-CMs through ATP stimulation of the mitochondrial ETC, which, in turn, reduced ROS-producing NOX4 activity
(A) Quantitative RT-PCR of NOX4 gene expression levels after the addition of NOX4-targeted Antisense LNA GapmeRs to the DMD hiPSC-CM cultures (left panel). As a positive control for the efficiency of the Antisense LNA GapmeRs, MALAT1 levels were determined after the addition of MALAT1-targeted Antisense LNA GapmeRs (right panel). Each data point is represented as ΔCt and is normalized for the housekeeping genes (GAPDH and RPL13a). Data are representative of three independent experiments (n = 3), and values are expressed as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
(B) Quantification of the NOX4 ROS production, measured via the NADPH-dependent ROS generation, in the isolated mitochondrial fraction of DMD hiPSC-CMs after a 6-day preincubation with GapmeRs. Each data point is represented as a percentage (%) and is normalized to the mitochondrial fraction of the untreated DMD hiPSC-CMs (vehicle).
(C) Quantification of the NADPH-dependent ROS production of NOX4 in the mitochondrial fraction of DMD hiPSC-CMs with or without idebenone treatment compared with in DMD isogenic and healthy controls.
(D) ATP luminescence detection showing the effect of idebenone treatment on the mitochondrial ATP levels in DMD hiPSC-CMs.
(E) Quantification of the ROS-producing NOX4 activity after 2.5 mM ATP addition in DMD hiPSC-CM and control cultures. Each data point is represented as a percentage (%) and is normalized to the mitochondrial fraction of the untreated DMD hiPSC-CMs.
(F) Quantification of the NADPH-dependent ROS production of NOX4 in the mitochondrial fraction of DMD hiPSC-CMs upon 2.5 mM ATP addition with or without idebenone treatment. Each data point is represented as a percentage (%) and is normalized to the mitochondrial fraction of the idebenone-treated DMD hiPSC-CM cultures. Data are representative of four or six independent experiments (n = 4 or n = 6), and values are expressed as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Colored rectangles represent the independent experiments.
See also Figures S7A–S7H.