Figure 4.
miR-302/367 and EEA motif act synergistically to promote pluripotency at the mid-reprogramming stage
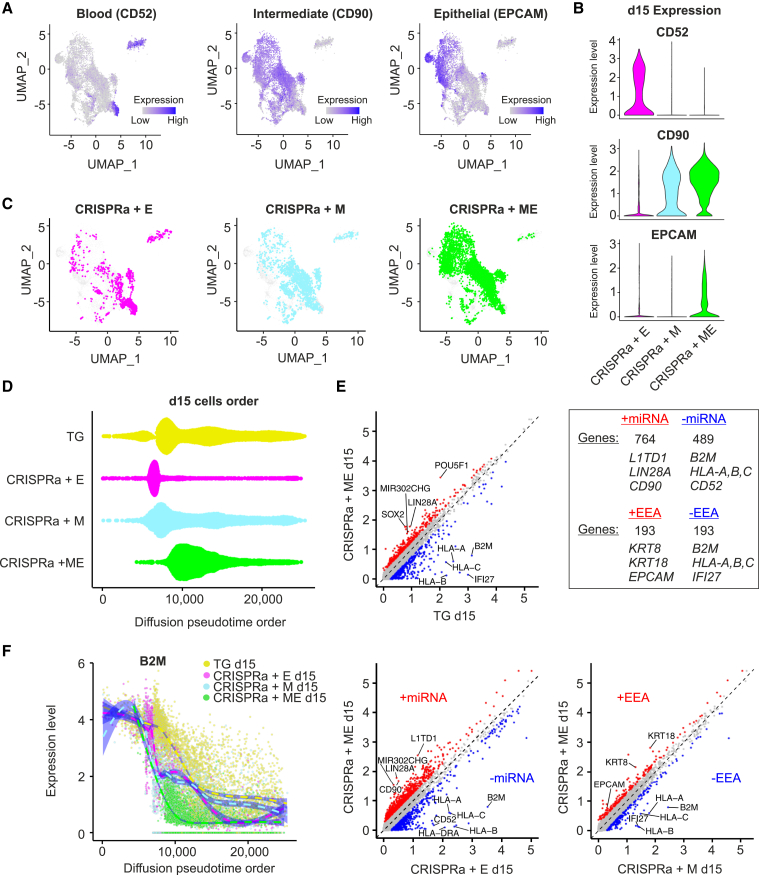
(A) Expression of blood cell marker CD52, intermediate reprogramming marker CD90, and epithelial marker EPCAM in all mid-reprogramming cells. Colors indicate expression levels (blue, high; gray, low).
(B) Violin plots showing the expression levels of CD52, CD90, and EPCAM across the different CRISPRa reprogramming conditions. See also Figure S4H.
(C) UMAPs showing the distribution of mid-reprogramming CRISPRa + E, CRISPRa + M, and CRISPRa + ME cells. Gray dots represent cells from other mid-reprogramming cell samples.
(D) Mid-reprogramming cells ordered by diffusion pseudotime. See also Figure S4B.
(E) Comparison of expression profiles between mid-reprogramming CRISPRa + ME and TG cells (top left), CRISPRa + ME and CRISPRa + E (bottom left), and CRISPRa + ME and CRISPRa + M (bottom right). Log-scaled average expression levels of all cells in each condition are plotted. At top right, the number of significantly highly expressed genes in CRISPRa conditions with or without miR-302/367 or EEA targeting are shown, with some of the top hits listed. These genes are marked as red or blue in the plots.
(F) Diffusion pseudotime combined with the expression level of B2M in the mid-reprogramming samples. Dashed lines and blue shades are fitted generalized additive model, with 95% confidence interval in each condition. Each color represents the different reprogramming condition.