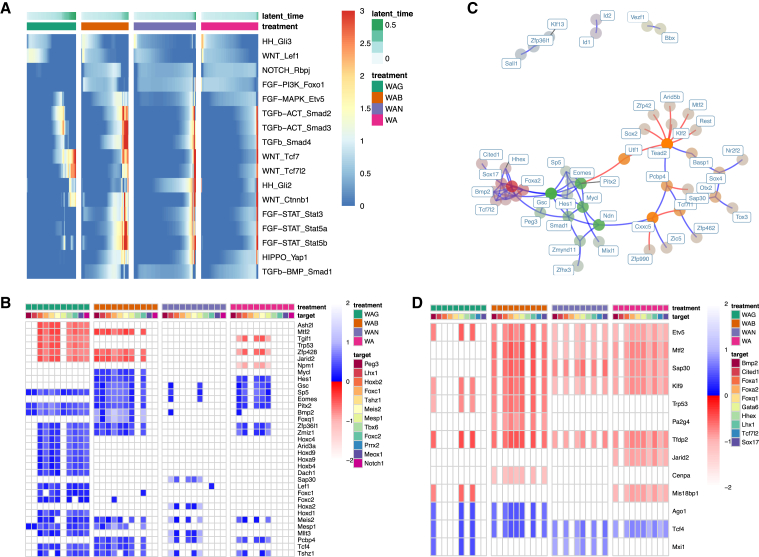
Figure 5.
Integrating signaling effectors and tracing paths to target genes
(A) Average expression of targets of signaling effectors by treatment over time. Pathway and effector are labeled on each row.
(B) Shortest path analysis showing length of shortest paths from Wnt effector targets (each row) to mesodermal target genes (each column) within treatment-specific networks. Path lengths are normalized against average path length within a network. Blue and red indicate paths that activate and repress the target gene, respectively. If such a path does not exist, then it has length of infinity and is therefore white.
(C) The epoch 3 subnetwork of the endodermal dynamic network. TFs are colored by community and faded by betweenness. Blue and red edges represent activating and repressive edges, respectively.
(D) Shortest path analysis showing length of shortest paths from PI3K suppression (targets of Foxo1 activation) (each row) to endodermal genes (each column) within treatment-specific networks. As before, path lengths are normalized against average path length within a network. Blue and red indicate activation and repression of the target gene, respectively. If a path does not exist, it has length of infinity and is therefore white.