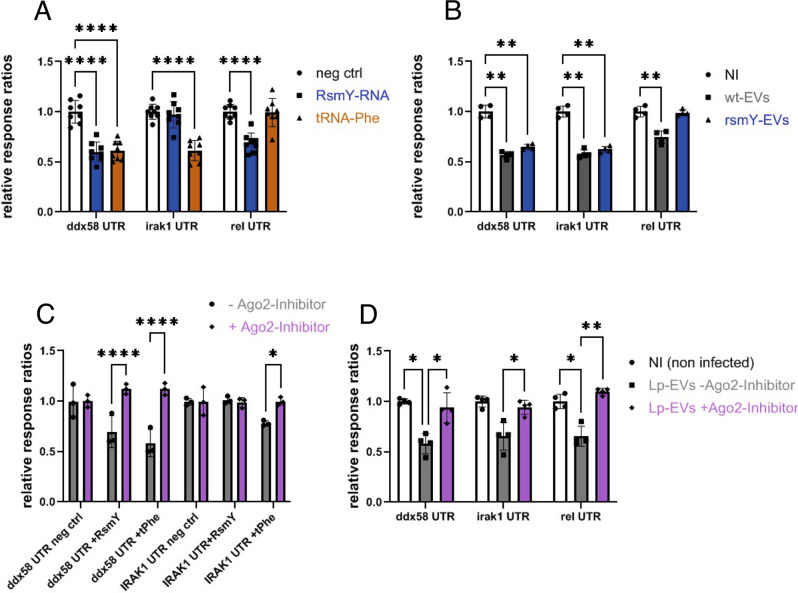
Fig. 4. RsmY base pairs with the UTR of RIG-I and RsmY activity is modulated by Ago2 inhibition.
A Dual luciferase reporter assay showing luciferase activities of THP-1 cells transfected with random short Lp-RNA (ctrlRNA), RsmY, or tRNA-Phe. Results are shown as relative luciferase activity (ratio Firefly/Renilla activity), where luciferase activity of ctrlRNA is defined as 100%. Bars indicate the (mean) SD of eight independent experiments. For statistical analysis, a two-way ANOVA was performed with p < 0.0001 (****). Source data provided as Source data file. B Dual luciferase reporter assay showing luciferase activities of THP-1 cells in absence (NI) or presence of wt or a ∆rsmY Lp-EVs. Lp-EVs affect protein expression depending on the presence of the un-translated regions (UTR) of ddx58 (RIG-I), irak1, or rel (cRel). Results are shown as relative luciferase activity (ratio Firefly/Renilla activity), luciferase activity of THP-1 cells (NI) was defined as 100%. Bars show the (mean) SD of n = 7 independent experiments. Statistical analysis, a two-way ANOVA with p < 0.05 significant (*), p < 0.01 very significant (**) and p < 0.001 extremely significant (***) was performed. Source data provided as Source data file. C Relative luciferase activities of THP-1 cells transfected with RsmY or tRNA-Phe. RsmY and tRNA-Phe affect protein expression significantly less in presence of an Ago2-Inhibitor. The bars indicate the (mean) SD of three independent experiments. Statistical analysis, a two-way ANOVA analysis was performed with p < 0.0001 (****). Source data provided as Source data file. D Dual luciferase reporter assay showing the relative luciferase activities (ratio Firefly/Renilla activity) of THP-1 cells incubated with Lp-EVs+/− Ago2-Inhibitor. When inhibiting Ago2, the suppressing effect of Lp-EVs on relative luciferase activity depending on the presence of the un-translated regions (UTR) of ddx58 (RIG-I), irak1 or rel (cRel) is significantly reduced. The bars show the (mean) SD of n = 4 independent experiments. Statistical analysis, a two-way ANOVA with values of p < 0.05 significant (*), p < 0.01 very significant (**) and p < 0.001 extremely significant (***) and p < 0.0001 (****) was performed. Source data provided as Source data file.