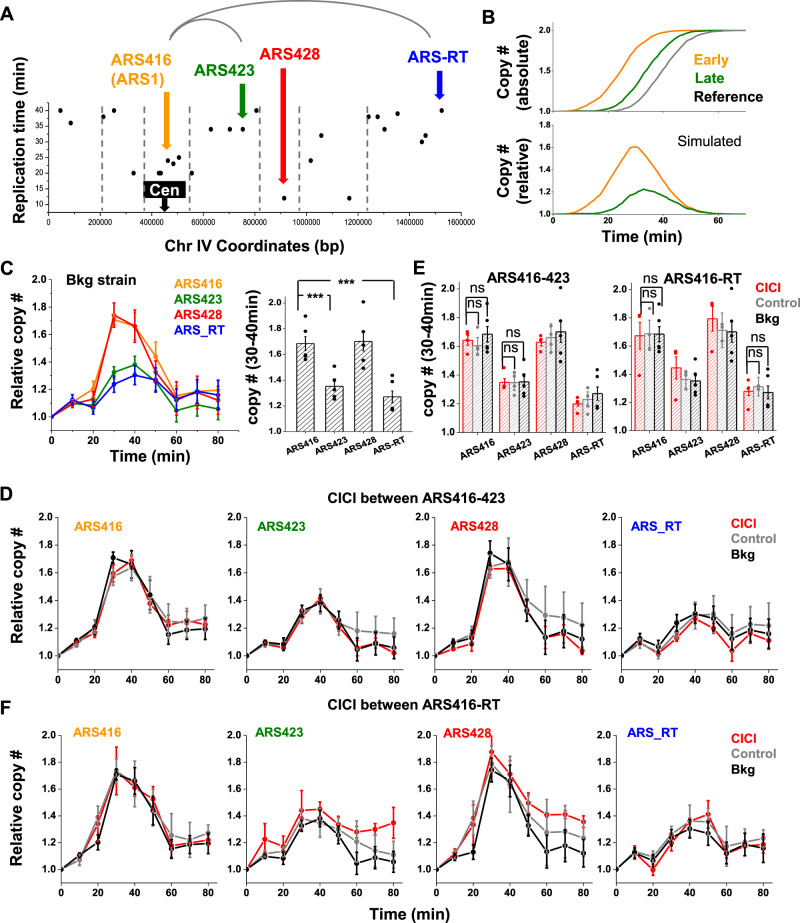
Fig. 6. CICI has no effect on replication timing.
A The replication timing of different ARS sequences (data from Raghuraman et al.37). The vertical lines represent TAD boundaries48. We choose four ARSs in four different TADs, ARS416, ARS423, ARS428, and ARS-RT for further testing. B Expected copy number as a function of time for different ARS sequences, as well as the relative copy number normalized by a control region with late replication. C The relative copy number as a function of time of the ARS sequences highlighted in A. Five biological repeats were included. The bar diagram on the right shows the average copy number of each ARS between 30 and 40 min after G1 release. Error bars represent standard error (same for panels below). The differences between ARS416 and ARS423/ARS-RT are statistically significant (one-sided Student’s t-test, P values < 0.001). D Consequence of CICI between ARS416 and ARS423 on replication timing (four biological repeats for each of CICI and control). The four panels show the relative copy number of the four ARSs in the CICI strain (LacO and TetO inserted near ARS416 and ARS423), control strain (same LacO and TetO arrays, but LacI and TetR are not conjugated to FKBP12 and FRB), and background strain (no CICI components). All the measurements were carried out in the presence of rapamycin. E Average ARS copy number between 30 and 40 min derived from data in D and F. No significant difference in replication timing was detected in CICI condition (two-sided Student’s t-test). F Same as in D except the CICI is between ARS416 and ARS-RT (TetO inserted near ARS-RT in the CICI and control strains, three biological repeats for each of CICI and control). Source data for B–F are provided as a Source Data file.