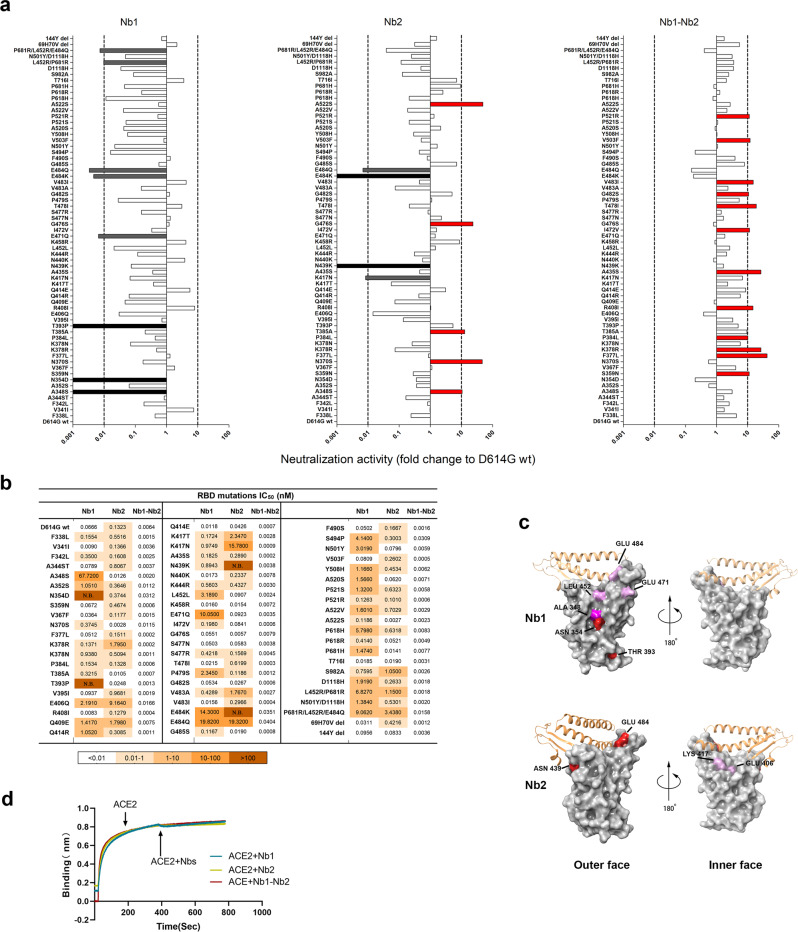
Fig. 3.
Epitope mapping using naturally occurring Spike mutants. a The SARS-CoV-2 pseudoviruses were packaged using more than 60 Spike variants identified from circulating viral sequences. The majority of mutations occur on RBD, including single amino acid substitution, combinational mutation and deletion. Neutralization activity conferred by Nb1, Nb2, and bivalent Nb1–Nb2 was evaluated. The x axis shows the ratio of IC50 of D614G pseudovirus/IC50 of indicated pseudovirus variant. When the ratio is greater than 1, the neutralization activity is increased, otherwise, the activity is decreased. The y axis shows the names of mutations. Data are represented as mean. All experiments were repeated at least twice. b IC50 values of indicated Nbs against SARS-CoV-2 mutation pseudovirus were calculated from data in (a). c Location of critical amino acids on the RBD (PDB ID: 6M0J) region for Nb1 and Nb2. The key hot spots targeted by Nbs are shown in a color-coding pattern with resistant strength descending from red to pink. Both sides of RBD are shown from different angles. d Competition between Nbs and ACE2 for binding to the SARS-CoV-2 RBD. Octet sensors immobilized with the SARS-CoV-2 RBD were first saturated with ACE2 protein and then exposed to the Nb1, Nb2, or Nb1–Nb2. The experiments were independently performed twice, and similar results were obtained