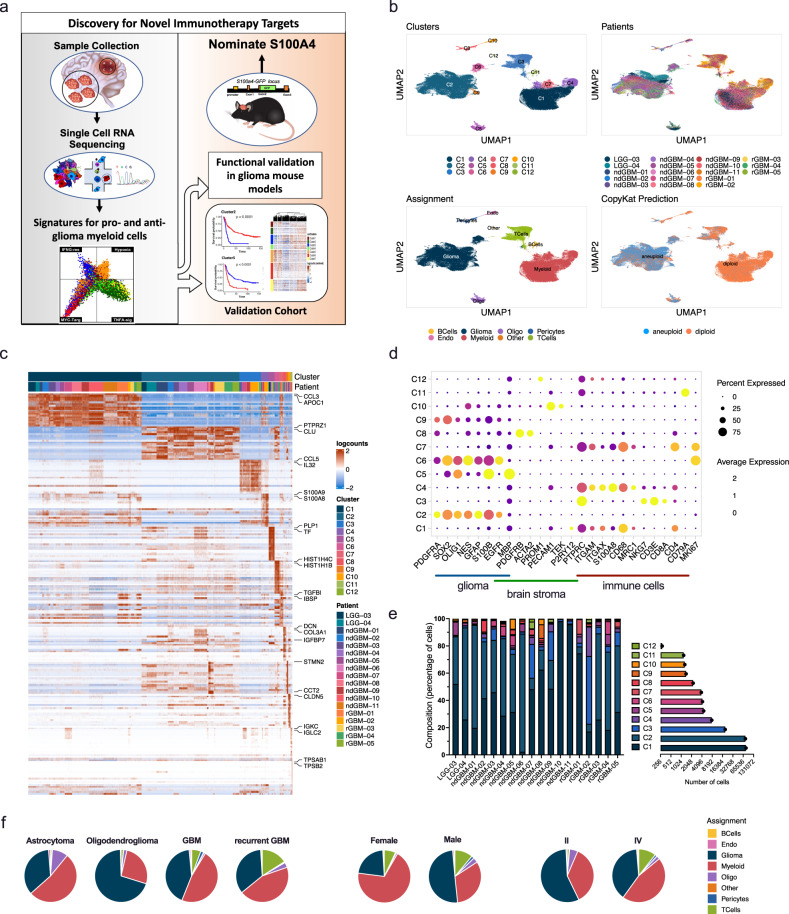
Fig. 1. Single-cell transcriptome analysis of human glioma and immune cells.
a A schematic summary of the study design. b UMAP projections of 201,986 aggregate single cells from 18 patients showing the composition of different cell types in human gliomas. UMAP projections are shown by cluster numbers, by the patient, by cluster assignment, and by diploid(normal)/aneuploid(malignant) status determined by CopyKat analysis (see Supplementary Fig. 2a). c Top 20 differentially expressed genes in clusters, ranked by FDR, are shown in the heatmap. Gene expression values were centered, scaled, and transformed to a scale from −2 to 2. Select signature genes are highlighted on the right. d Dot plot showing marker gene expression for different cell types (gliomas, brain stroma (pericytes and oligodendrocytes), and immune cells). Dot sizes indicate the percentage of cells in each cluster expressing the gene, and colors indicate average expression levels. e Fraction of cells (y-axis) from each patient sample (x-axis) color-coded for cluster IDs as in (b, c). The numbers of cells in each cluster from all patients are also indicated in the horizontal bar graph on the right. (Also see to Supplementary Data 2). f Pie charts representing the percentage of cells per assignment by tumor type, sex, and tumor grade color-coded for cell type assignment. Source data for e and f are provided as a Source Data file.