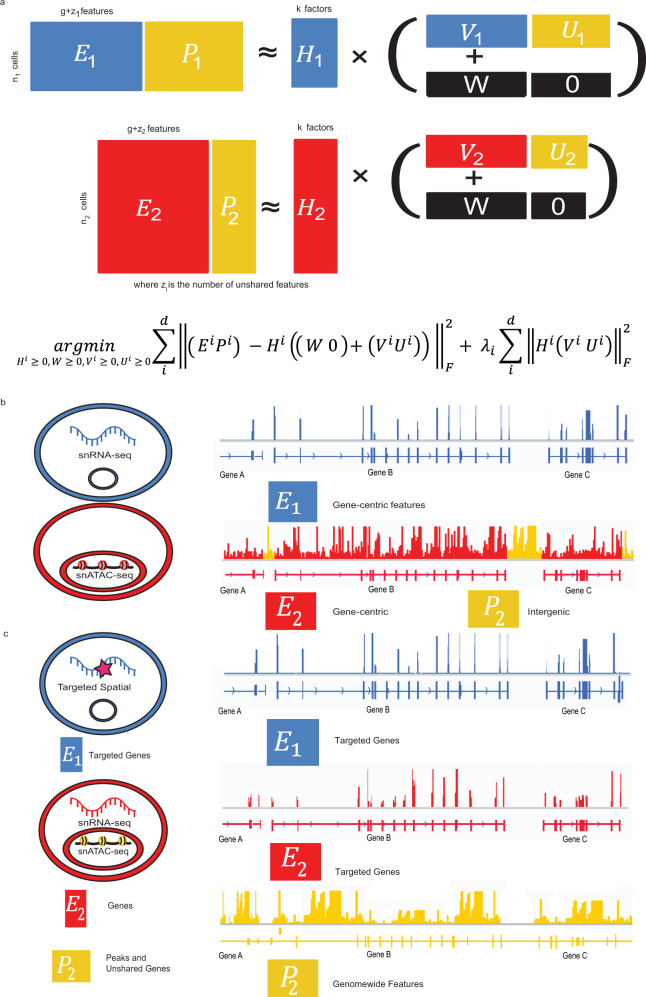
Fig. 1. Overview of the UINMF algorithm for integrating single-cell datasets with partially overlapping features.
a Schematic representation of the matrix factorization strategy (top) and optimization problem formulation (bottom). The addition of a factor matrix Ui allows for unshared features to be utilized in joint matrix factorization. Each dataset (Ei) is decomposed into shared metagenes (W), dataset-specific metagenes constructed from shared features (Vi), unshared metagenes (Ui), and cell factor loadings (Hi). The incorporation of the U matrix allows unshared features that occur in only one dataset to inform the resulting integration. b UINMF can integrate data types such as scRNA-seq and snATAC-seq using both gene-centric features and intergenic information. c UINMF can integrate targeted spatial transcriptomic data with simultaneous single-cell RNA and chromatin accessibility measurements using both unshared epigenomic information and unshared genes.