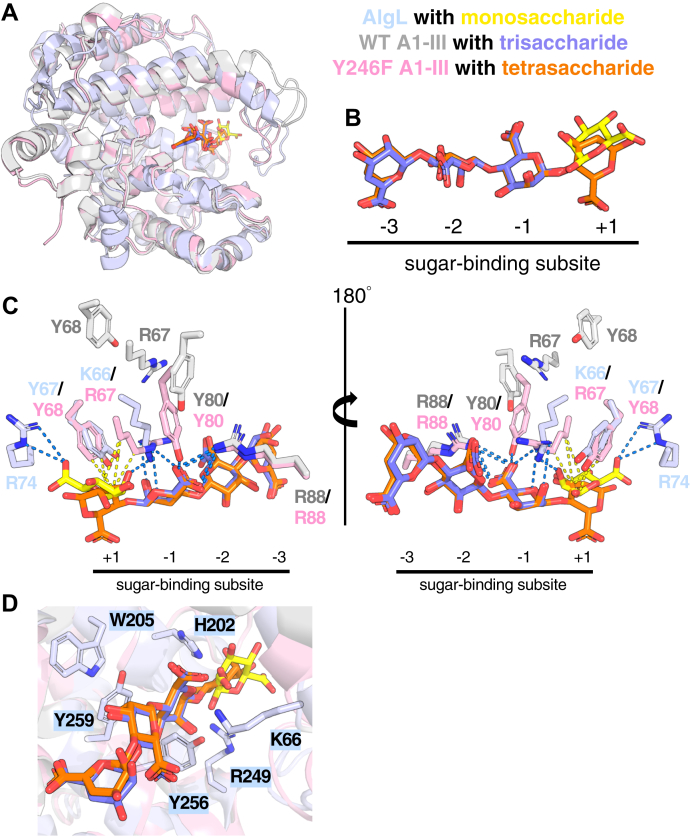
Figure 3.
The polysaccharide family 5 bacterial alginate lyases Pseudomonas aeruginosa AlgL and Sphingomonas sp. A1-III are structurally similar.A, superposition of P. aeruginosa AlgL (light blue) in complex with a monosaccharide of mannuronate (yellow) (PDB: 4OZV), Sphingomonas sp. WT A1-III (grey) in complex with a mannuronate trisaccharide (purple) (PDB: 1HV6) (52), and Sphingomonas sp. Y256F A1-III (pink) in complex with a mannuronate tetrasaccharide (orange) (PDB: 4F13) (53). B, ligands bound and their sugar-binding subsite positions in the P. aeruginosa AlgL and Sphingomonas sp. A1-III structures. C, close-up of the lid-loop region of the P. aeruginosa AlgL and Sphingomonas sp. A1-III active sites. Hydrophobic and hydrogen-bonding interactions are represented by the dashed yellow and blue lines, respectively. D, P. aeruginosa AlgL active site residues (light blue) chosen for downstream in vitro and in vivo mutagenic studies, including the proposed catalytic acid and base Y256, the residue that stabilizes the anionic intermediate H202, the lid-loop residue K66, the highly conserved residues W205 and Y259, and R249 involved in neutralizing the C-5 carboxylate group.