Figure 1.
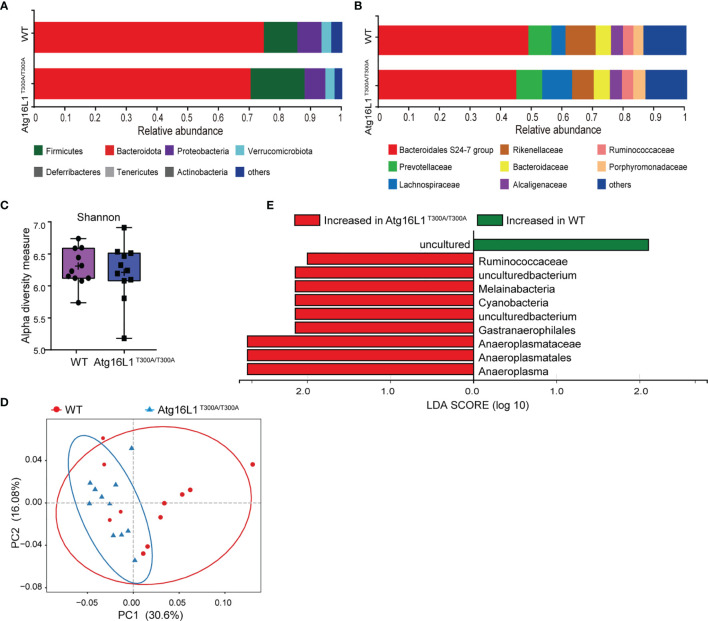
Atg16L1T300A mice display a slightly different microbial composition in fecal at a steady state. WT and Atg16L1T300A/T300A mice were cohoused in a specific-pathogen-free (SPF) facility for 4–5 weeks after weaning at the third week of birth. (WT mice n = 11 and Atg16L1T300A/T300A mice n = 11). Stool samples were collected and analyzed by 16S rRNA gene amplicon sequencing. (A, B) The relative abundance of dominant bacteria at phyla (A) and families (B) level microbiota composition determined by 16S rRNA gene amplicon sequencing of stool samples from SPF cohoused WT vs. Atg16L1T300A/T300A mice at steady state. Colored bars represent the relative abundance of particular taxa averaged in each group. (C) Box-and-whisker plot (boxes show median, whisker denotes minimum to maximum range) of microbiota alpha-diversity within the fecal microbiomes of WT vs. Atg16L1T300A/T300A SPF mouse samples calculated using the Shannon index. (D) PCoA plot of the fecal microbiota composition (unweighted UniFrac distances). (E) Histogram of LDA value distribution identified by LEfSe analysis. Each symbol represents an individual animal (non-parametric test followed by a Mann–Whitney test).