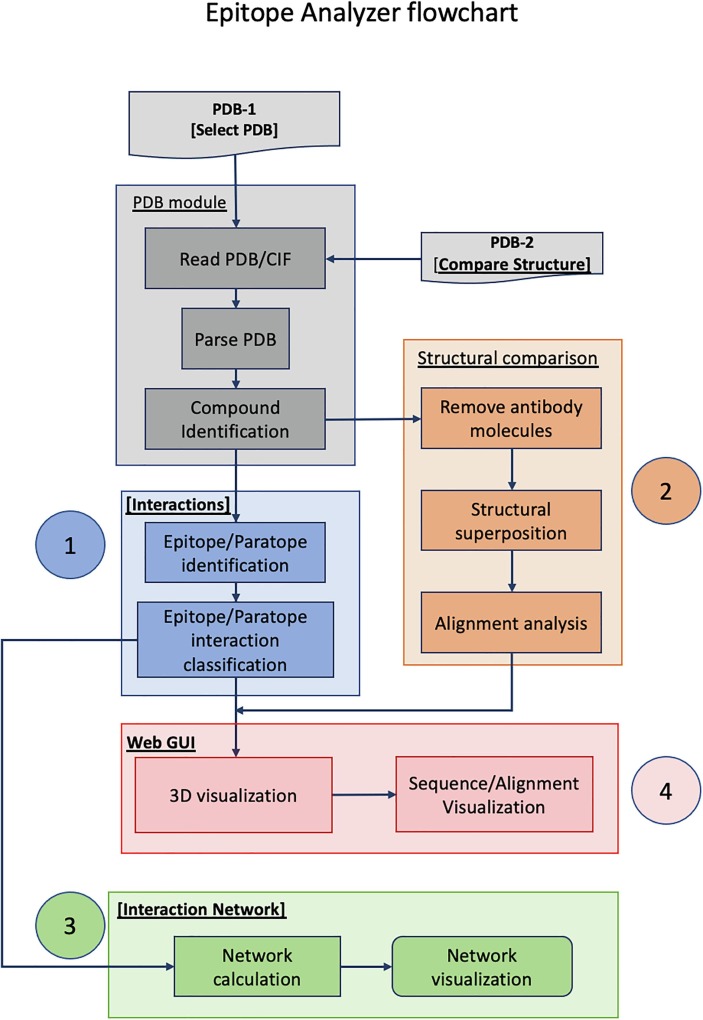
Fig. 1.
Flowchart description of Epitope Analyzer (EA) tool (http://viperdb.scripps.edu/ea.php). There are 4 main components in the EA tool that overlap with each other: 1) Ag-Ab interaction analysis, 2) analysis of epitope conservation, 3) network analysis and 4) graphical user interface (GUI), identified blue, orange, green and pink colored areas. PDB-1 is the reference structure file of an antigen–antibody complex that can either be downloaded directly from PDB by specifying PDB-ID (e.g., 3gbn) or uploaded by the user from his/her computer. The molecular entities of PDB-1 and their sequences are displayed in the Web-GUI window. The [interactions] module calculates the non-covalent interactions between the selected chains using a chosen distance cutoff. The identified epitope and paratope residues are listed in one of the side panels and displayed on demand in the Web-GUI window. The antigen structure to be compared (PDB-2) is uploaded in [Structural comparison] panel. The results of the structure comparison and similarity in epitope residues are displayed in one of side the panels. The [Interaction Network] panel calculates and displays the network of interactions between the selected chains in the reference structure (PDB-1). Finally, the lists of epitope residues and sequence alignments can be downloaded in “CSV” and “FASTA” formats, respectively from the hyperlinks provided. JSMol is an open-source HTML-5 viewer for 3D molecular structures (http://wiki.jmol.org/index.php/JSmol).