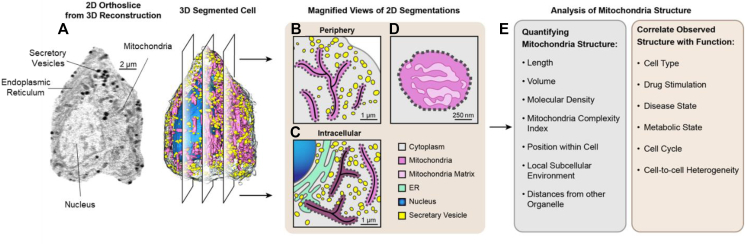
Figure 1.
Process of using SXT to correlate mitochondrial structure and function. Modified from [16]. a) Representative 2D orthoslice and a reconstruction of a β-cell (corresponding 3D map) display the segmented organelles. The segmented cell masks are then used to quantify specific features of the cellular organization such as mitochondrial network structure. b-c) Magnified view of 2D slices of the segmented cell illustrate features that can be extracted and compared between different cellular regions. SXT can reveal slight differences in biochemical compositions of organelles by close analysis of their LAC values. An illustration of mitochondria with different biochemical compositions based on cellular localizations is shown in panel c. The darker pink shade indicates denser mitochondria. d) Illustration of a cross section of a mitochondria with cristae shown in dark pink and the mitochondrial matrix shown in light pink. The degree of mitochondrial complexity or extent of cristae folding can be quantified by segmenting these subcompartments using LAC values. e) List of the specific features that can be quantified from 3D segmentations of a cell and the corresponding cellular conditions that can be compared to correlate structure and function.