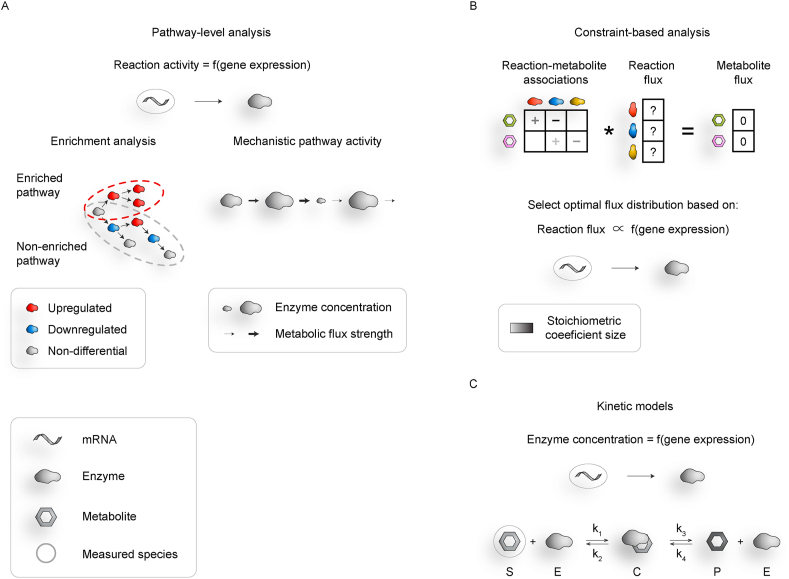
Figure 2.
Metabolic modelling approaches. (A) Pathway-level analysis assumes that gene expression maps directly to enzyme concentration or reaction activity. It can be used for defining pathways enriched in differentially expressed genes or for calculating the activity of individual pathways by propagating activity through the pathway (mechanistic pathway analysis). (B) Constraint-based analysis assumes that metabolite concentrations are constant and based on this defines possible fluxes that would result in such a system, as further described in the main text. Namely, metabolite fluxes, which are constrained to be zero, are obtained by multiplying reaction fluxes with the stoichiometric matrix that defines metabolite conversions involved in each reaction. This gives an undetermined system of equations that defines possible reaction flux distributions. Optimal flux configuration is then selected based on optimization objectives, such as correspondence between gene expression and enzyme activity. (C) Kinetic models predict metabolic changes based on detailed prior metabolic knowledge and information on metabolite and enzyme availability. Gene expression can be used for the prediction of enzyme concentration.