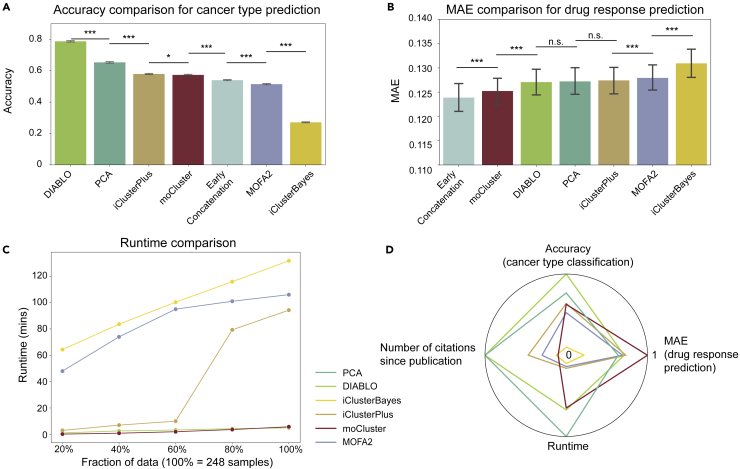
Figure 5.
Benchmarking of machine learning-based integration tools using the CCLE multi-omics data
(A) Accuracy of each method for cancer type prediction, showing standard errors of the mean derived from 100 runs of five-fold cross-validation, totalling 500 experiments (∗ signifies p value < 0.05 and ∗∗∗ signifies p value < 0.001 by an unpaired two-tailed Student’s t test).
(B) MAE comparison for drug response prediction across 1,448 drugs, error bars representing standard errors of the mean (∗∗∗ signifies p value < 0.001 and n.s. stands for not significant by an unpaired two-tailed Student’s t test).
(C) Runtime comparison. PCA is omitted as the runtime was negligible compared with the five multi-omics integration methods.
(D) A summary of the benchmarking study, derived from the results of cancer type prediction, drug response prediction (MAE between the measured AUC and predicted AUC), runtime comparison, and the number of citations since publication. The number of citations for PCA was set to the maximum for better visualization and because of its widespread use. The inverse of the runtime and drug response prediction MAE values are plotted so that higher values indicate better performance in all dimensions, and all values are plotted in the range of 0 to 1 in the radar plot.