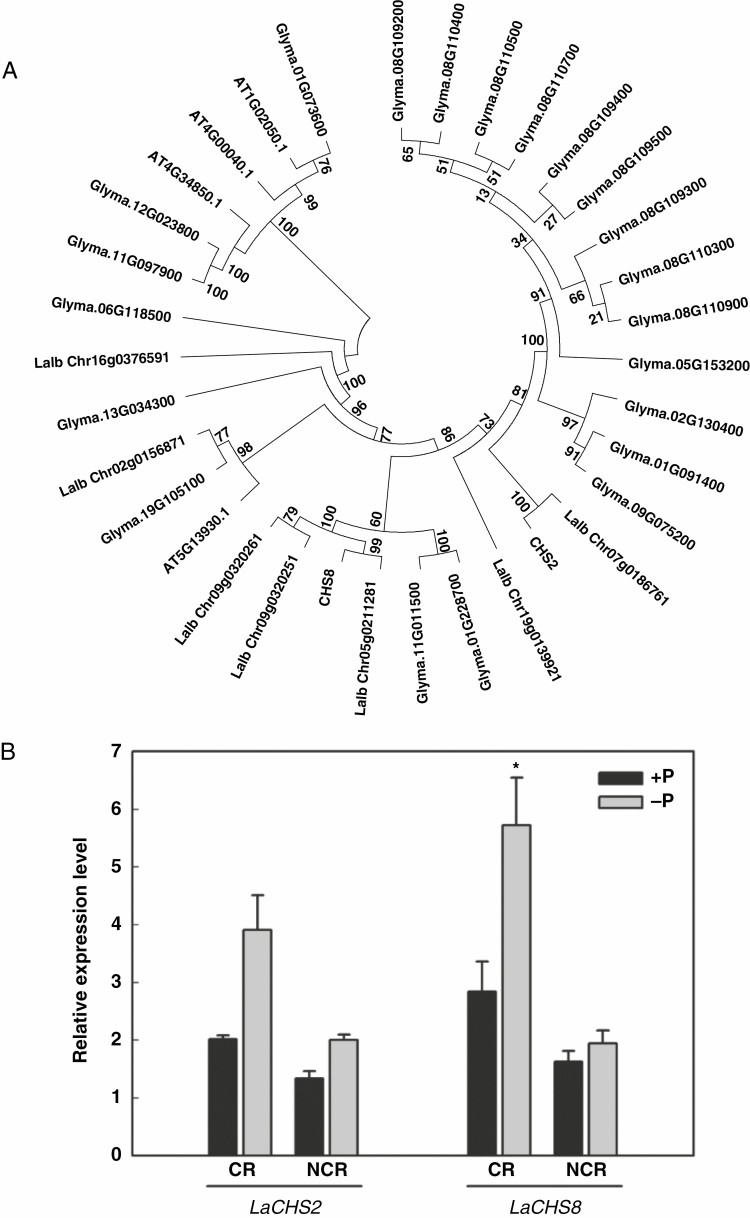
Fig. 2.
Phylogenetic tree based on sequences of CHS proteins and expression of LaCHS2 and LaCHS8 in roots of white lupin grown under P-sufficient and P-deficient conditions. (A) Sequences were derived from NCBI and WHITE LUPIN GENOME (https://www.whitelupin.fr/index.html). The alignments were saved and executed using MEGA to generate a neighbour-joining tree. (B) qRT–PCR analysis of CHS expression in different root types of white lupin grown under P-sufficient and P-deficient conditions. Plants were grown for 21 d with 75 μm P (+P) or 0 μm P (−P). Cluster roots (CR) and non-cluster roots (NCR) were harvested and used for RNA isolation and cDNA synthesis. qRT–PCR was used to assess LaCHS2 and LaCHS8 transcript abundance in response to P treatments. Data are expressed as relative values based on the LaCHS expression level in +P non-cluster roots. Asterisk in (B) indicates significant difference between −P and +P by Student’s t-test: *P < 0.05. Significant difference was based on four biological replicates. Bars on the graph are standard errors.