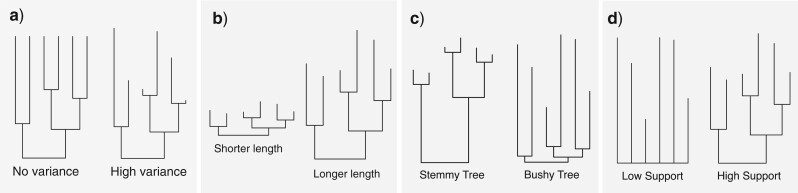
Figure 1.
Four branch statistics used to test branch-length signal in each gene tree: a) coefficient of variation in root-to-tip distances, which provides a measure of rate variation across lineages, or inaccuracies in branch-length estimation; b) total tree length (calculated as the sum of branch-lengths), which indicates the overall substitution rate at a locus; c) stemminess, defined as the ratio of internal to terminal branch lengths; and d) Shimodaira—Hasegawa-like approximate likelihood-ratio test (SH-aLRT) of mean branch supports, which can be taken as a measure of the consistency of the topology signal across alignment sites.