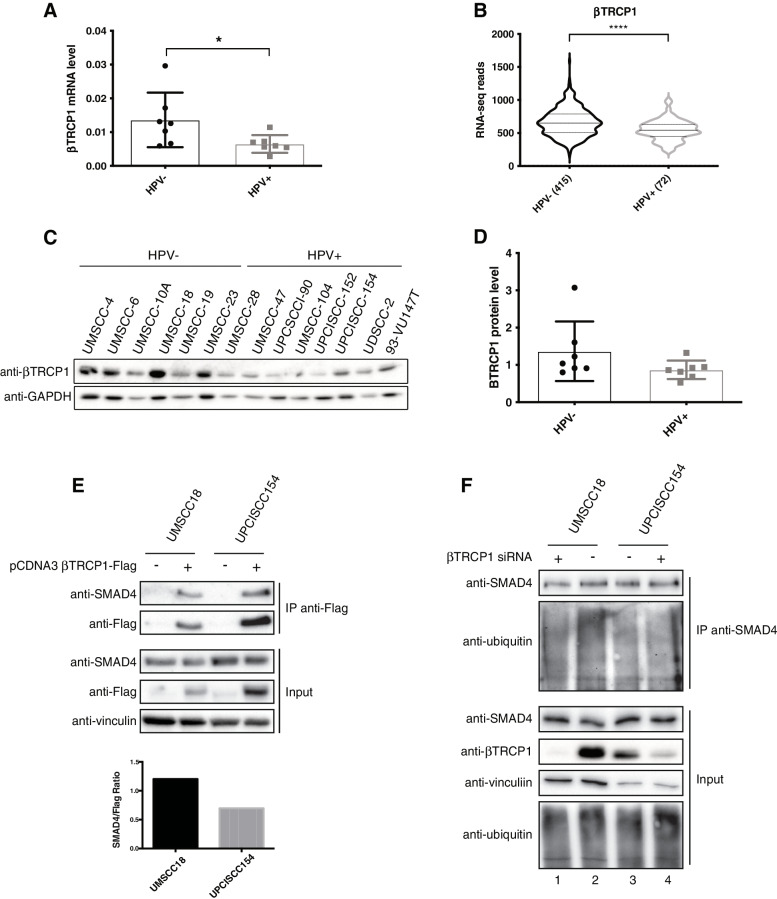
Fig. 4.
Analysis of βTRCP1-dependent SMAD4 degradation in HPV-negative and HPV-positive HNC cell lines. A Total RNAs from HNC cell lines were isolated for RT-qPCR. βTRCP1 expression was normalized to RpP0 and shown as Box plot of means ± SD and individual values of at least three independent experiments. *, P < 0.05 (unpaired t test). B Normalized RNA-seq data were extracted from TCGA database and divided in HPV+ and HPV- HNC cohorts. Numbers in brackets refer to the number of samples included in each analysis. ****, P < 0.0001 (unpaired t test). C HNC cell lines were lysed and analysed by immunoblotting with the indicated antibodies. D Box plot showing means ± SD and individual values of Optical density analysis of the expression of βTRCP1 normalised to loading control (GAPDH) from three independent western blot experiments in HNC cell lines. E pcDNA3-βTRCP1-flag plasmid was transfected into HPV-positive (UPCISCC154) and HPV-negative (UMSCC18) HNC cell lines. One day after transfection cell were treated with 5 μM MG132 overnight. Two days after transfection cells were harvested and whole cells extracts were subjected to anti-Flag immunoprecipitation (IP). Immunoblotting with the indicated antibodies was then performed and whole cell extracts were used as input controls. Graph shows Optical density analysis of SMAD4 co-immunoprecipitated normalised to βTRCP1-flag immunoprecipitated. F HPV-positive (UPCISCC154) and HPV-negative (UMSCC18) HNC cell lines were transfected with specific siRNA against βTRCP1 or Luciferase. Two days after transfection cell were treated with 5 μM MG132 overnight. 72 h after transfection cells were harvested and whole cells extracts were subjected to anti-SMAD4 immunoprecipitation (IP). Immunoblotting with the indicated antibodies was then performed and whole cell extracts were used as input controls