Table 1.
Chemical structure, name, docking or binding score, binding free energy, platform, and Protein Data Bank ID for best-reported drug candidates against the S protein of SARS-CoV-2.
| Entry no. | Compound | Docking/binding score (kcal/mol) | Binding free energy (MM-GBSA; kcal/mol) | Platform (Protein Data Bank ID) | Refs |
|---|---|---|---|---|---|
| 1 |
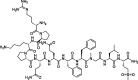 Sar9 Met (O2)11-Substance P |
– | −10.63 | Molecular operating environment (MOE) (6LZG) | 25 |
| 2 |
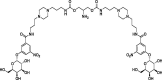 BV2 |
– | −17.32 | MOE (6LZG) | 25 |
| 3 |
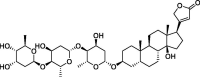 Digitoxin |
−8.7 | – | Autodock; AMBER16 (6LZG) | 26 |
| 4 |
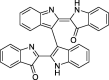 Bisindigotin |
−8.3 | – | Autodock; AMBER16 (6LZG) | 26 |
| 5 |
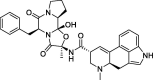 Ergotamine |
−8.8 | – | Autodock; (6ACD) | 27 |
| 6 |
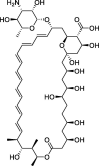 Amphotericin b |
−8.3 | – | Autodock; (6ACD) | 27 |
| 7 |
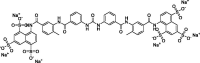 Lig8970 |
−8.7 | −40.43 | Autodock; GROMACS (5X5B, 6ACG, 5I08) | 28 |
| 8 |
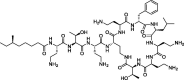 Polymyxin B |
−99.4 | −164.3 | PLANTS; AMBER (6M0J) | 29 |
