Table 2.
Chemical structure, name, docking or binding score, binding free energy, platform, and Protein Data Bank ID for best-reported drug candidates against the main proteases (Mpro or 3CLpro) of SARS-CoV-2.
| Entry No. | Compound | Docking/binding score (kcal/mol) | Binding free energy (MM-GBSA) (kcal/mol) | Platform (Protein Data Bank ID) | Refs |
|---|---|---|---|---|---|
| 1 |
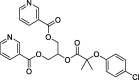 Binifibrate |
– | –69.04 | Schrodinger (6 W63) | 30 |
| 2 |
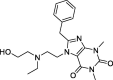 Bamifylline |
– | –63.19 | Schrodinger (6 W63) | 30 |
| 3 |
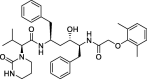 Lopinavir |
−9.9 | – | PyRx 0.8 (6Y84) | 31 |
| 4 |
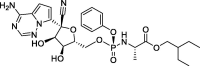 Remdesivir |
−9.7 | – | PyRx 0.8 (6Y84) | 31 |
| 5 |
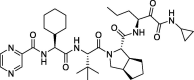 Telaprevir |
−10.05 | – | Autodock (6LU7) | 32 |
| 6 |
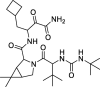 Boceprevir |
−9.15 | – | Autodock (6LU7) | 32 |
| 7 |
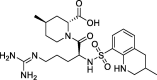 Argatroban |
−9.03 | – | Autodock (6LU7) | 32 |
| 8 |
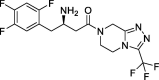 Sitagliptin |
−8.80 | – | Autodock (6LU7) | 32 |
| 9 |
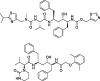 Lopinavir-Ritonavir |
−10.6 | – | Autodock (6Y2F) | 34 |
| 10 |
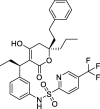 Tipranavir |
−8.7 | – | Autodock (6Y2F) | 34 |
| 11a |
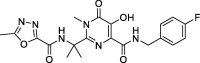 Raltegravir |
−8.3 | – | Autodock (6Y2F) | 34 |
| 12 |
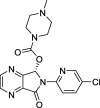 Eszopiclone |
−10.0 | –54.504 | Autodock (6Y2G) | 35 |
| 13b |
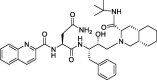 Saquinavir |
−9.1 | –125 | Autodock; GROMACS (6LU7) | 36 |
| 14b |
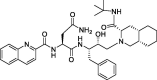 Saquinavir |
−9.5 | – | Autodock (6LU7) | 27 |
| 15 |
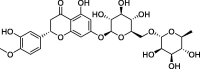 Hesperidin |
−8.3 | – | Autodock (6LU7) | 37 |
| 16 |
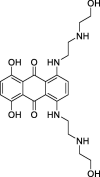 Mitoxantrone |
− | –43.5854 | Schrodinger (6LU7) | 38 |
| 17a |
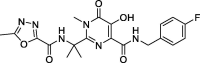 Raltegravir |
−9.0 | – | Autodock (6LU7) | 39 |
| 18 |
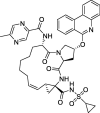 Paritaprevir |
−10.9 | – | Autodock (6Y2E) | 40 |
Chosen as top candidate in two studies.
Chosen as top candidate in two studies.
