Figure 5. Macrophages and IL-1β suppress the binding of PPARα to ENI and FOXO3 to ENII.
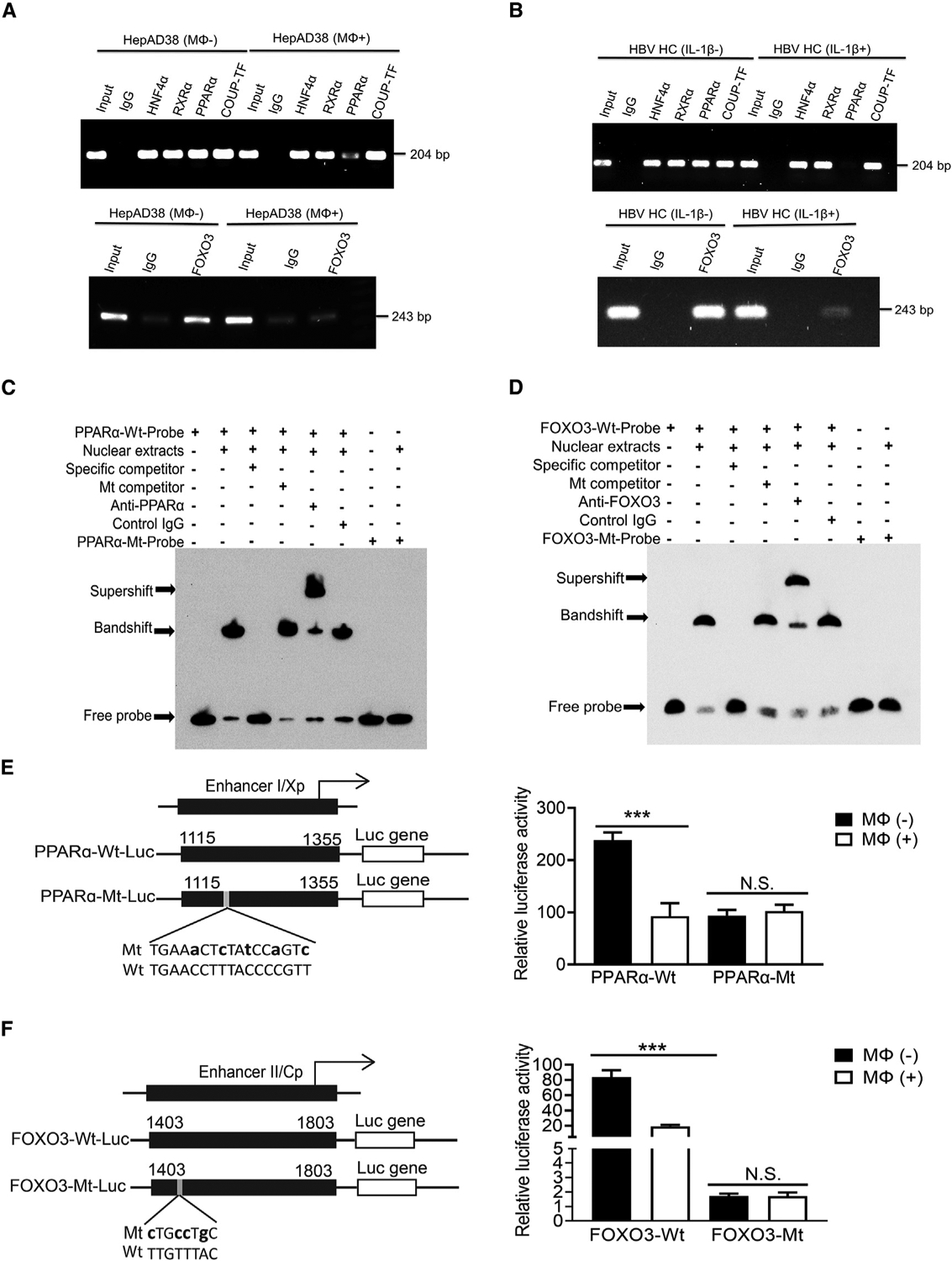
(A) The binding of HNF4α, RXRα, PPARα, and COUP-TF1 to the ENI enhancer (top panel) or FOXO3 to the ENII enhancer (bottom panel) was analyzed by the ChIP assay using HepAD38 cells with (Mφ+) or without (Mφ−) co-culturing with THP-1 macrophages. The control IgG served as the negative control. “Input”, total cell lysates without the step of immunoprecipitation to serve as the positive control.
(B) The same ChIP assay as shown in (A) was conducted using HCs isolated from HBV transgenic mice with (IL-1β+) or without (IL-1β–) the injection of IL-1β.
(C) The binding of PPARα to nt 1,136–1168 of ENI1 was analyzed by the EMSA. The double-stranded oligonucleotide containing the sequence of nt 1,136–1,168 was used as the probe for incubation with the HepG2 nuclear extracts. The nucleotide mutations of the mutant probe are shown in (E).
(D) The binding of FOXO3 to nt 1,403–1,455 of ENII was analyzed by the EMSA. The nucleotide mutations of the mutant probe are shown in (F).
(E) Wild-type (WT) and mutant (Mt) ENI/Xp Luc reporter constructs are shown to the left. The lowercase letters indicate the nucleotide mutations introduced. These reporter constructs were transfected into HepAD38 cells with or without co-culturing with THP-1 macrophages. The Luc reporter results are shown to the right.
(F) WT and Mt ENII/Cp Luc reporter constructs are shown to the left. The study was conducted as shown in (E). N.S., not significant; ***p < 0.001.
