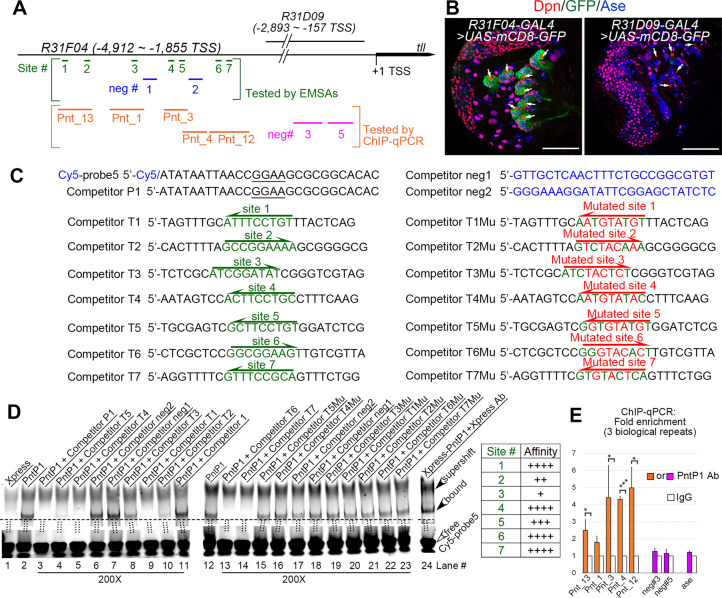
Fig 5. PntP1 directly binds to the tll enhancer in type II NBs.
(A) A diagram of locations of putative PntP1 binding sites or negative control sites in the tll enhancer regions of R31F04 or R31D09. Green lines and blue lines within square brackets indicate the PntP1 binding sites (sites #1–7) and the sequences (neg#1–2) lacking PntP1 binding sites, respectively, that are tested by EMSAs. Orange lines and magenta lines within the orange square brackets indicates DNA fragments with (Pnt_13, Pnt_1, Pnt_3, Pnt_4, Pnt_12) or without (neg#3, neg#5) corresponding PntP1 binding sites, respectively, that are tested by ChIP-qPCR. (B) GAL4 under the control of the tll enhancer subfragment R31F04 but not R31D09 specifically activates mCD8-GFP expression in type II NB lineages. White arrows point to type II NBs. Scale bars equal 50μm. (C) Sequences of competitors and probes used for EMSAs. Nucleotides in green indicate sequences of putative PntP1 binding motifs and nucleotides in red indicated mutations introduced into the putative PntP1 binding motifs. Underlined nucleotides in the Cy5-probe5 and the competitor P1 indicates the 5’-GGAA core sequence of the PntP1 binding motif. The direction of arrows indicates the orientations of the consensus 5’-GGAA/T-3’ core sequences. (D) EMSAs of the binding of Xpress-tagged PntP1 with Cy5-probe5 that contains a PntP1 binding motif from the erm enhancer fragment R9D11 and its competition by indicated competitors that contains corresponding wild type or mutated putative PntP1 binding motifs. Specific binding between Xpress-PntP1 and Cy5-probe5 is detected in lanes #2 and #12 and is verified by the presence of a super-shifted band in the presence of the Xpress antibody (lane #24) and the competition by the cold probe (competitor P1) (lane #3). Competitors that contain the wild type PntP1 binding sites (competitors T1-T7, corresponding to lanes #4–5, #8–10, and #13–14) can all compete with the Cy5-probe5 for the binding with Xpress-PntP1 to a various of degrees, but the competitors that contain mutated PntP1 binding motifs (competitors T1Mu-T7Mu, corresponding to lanes #15–16, and #19–23) or no PntP1 binding motifs (competitors neg1 and neg2 or the competitor 1 that contains the sequence from the kr enhancer, corresponding to lanes #6–7, #11, and #17–18) cannot. The relative binding affinity of the seven putative PntP1 binding site is summarized in the table to the right. Open arrowheads point to free probes while filled arrowheads point to the bands of PntP1-Cy5-probe5 complexes or the super-shifted band of the Xpress Ab-PntP1-Cy5-probe5 complex. The ratio of the competitor to the probe is 200 for all competitors tested. (E) DNA fragments (Pnt_13, Pnt_1, Pnt_3, Pnt_4, Pnt_12) that contain corresponding predicted PntP1 binding sites are all pulled down by the anti-PntP1 antibody from larval brains in the ChIP-qPCR assays, but not the DNA fragments neg#3 or neg#5 from the R31D09 enhancer region or a DNA fragment from the ase enhancer that do not contain putative PntP1 binding sites. The quantification data represent the Mean ± SD of the fold changes normalized to IgG controls of three biological replicates. *, P < 0.05. ***, P < 0.001.